| Motif | ZSC29.H12INVITRO.1.M.C |
| Gene (human) | ZSCAN29 (GeneCards) |
| Gene synonyms (human) | ZNF690 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 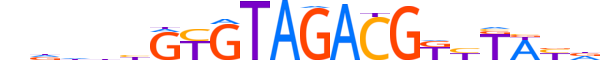 |
| Motif subtype | 1 |
| Quality | C |
| Motif | ZSC29.H12INVITRO.1.M.C |
| Gene (human) | ZSCAN29 (GeneCards) |
| Gene synonyms (human) | ZNF690 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 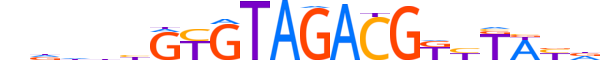 |
| Motif subtype | 1 |
| Quality | C |
| Motif length | 20 |
| Consensus | hRWARMCGTCTACACvdhhn |
| GC content | 44.34% |
| Information content (bits; total / per base) | 19.008 / 0.95 |
| Data sources | Methyl-HT-SELEX |
| Aligned words | 770 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 4 (25) | 0.648 | 0.686 | 0.544 | 0.621 | 0.666 | 0.688 | 1.997 | 2.406 | 39.161 | 63.745 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Methyl HT-SELEX, 1 experiments | median | 0.921 | 0.911 | 0.749 | 0.757 | 0.627 | 0.654 |
| best | 0.921 | 0.911 | 0.749 | 0.757 | 0.627 | 0.654 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | ZNF500-like {2.3.3.19} (TFClass) |
| TFClass ID | TFClass: 2.3.3.19.4 |
| HGNC | HGNC:26673 |
| MGI | |
| EntrezGene (human) | GeneID:146050 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZSC29_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q8IWY8 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 4 human, 0 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 1 |
| PCM | ZSC29.H12INVITRO.1.M.C.pcm |
| PWM | ZSC29.H12INVITRO.1.M.C.pwm |
| PFM | ZSC29.H12INVITRO.1.M.C.pfm |
| Alignment | ZSC29.H12INVITRO.1.M.C.words.tsv |
| Threshold to P-value map | ZSC29.H12INVITRO.1.M.C.thr |
| Motif in other formats | |
| JASPAR format | ZSC29.H12INVITRO.1.M.C_jaspar_format.txt |
| MEME format | ZSC29.H12INVITRO.1.M.C_meme_format.meme |
| Transfac format | ZSC29.H12INVITRO.1.M.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 103.5 | 261.5 | 51.5 | 353.5 |
| 02 | 430.0 | 73.0 | 213.0 | 54.0 |
| 03 | 94.0 | 56.0 | 31.0 | 589.0 |
| 04 | 635.0 | 61.0 | 37.0 | 37.0 |
| 05 | 380.0 | 52.0 | 291.0 | 47.0 |
| 06 | 482.0 | 225.0 | 20.0 | 43.0 |
| 07 | 2.0 | 754.0 | 3.0 | 11.0 |
| 08 | 69.0 | 0.0 | 701.0 | 0.0 |
| 09 | 0.0 | 0.0 | 12.0 | 758.0 |
| 10 | 0.0 | 743.0 | 0.0 | 27.0 |
| 11 | 0.0 | 0.0 | 0.0 | 770.0 |
| 12 | 768.0 | 0.0 | 0.0 | 2.0 |
| 13 | 6.0 | 665.0 | 1.0 | 98.0 |
| 14 | 476.0 | 1.0 | 292.0 | 1.0 |
| 15 | 41.0 | 639.0 | 12.0 | 78.0 |
| 16 | 344.0 | 108.0 | 220.0 | 98.0 |
| 17 | 314.0 | 109.0 | 175.0 | 172.0 |
| 18 | 276.5 | 134.5 | 107.5 | 251.5 |
| 19 | 158.75 | 361.75 | 68.75 | 180.75 |
| 20 | 237.75 | 222.75 | 125.75 | 183.75 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.134 | 0.34 | 0.067 | 0.459 |
| 02 | 0.558 | 0.095 | 0.277 | 0.07 |
| 03 | 0.122 | 0.073 | 0.04 | 0.765 |
| 04 | 0.825 | 0.079 | 0.048 | 0.048 |
| 05 | 0.494 | 0.068 | 0.378 | 0.061 |
| 06 | 0.626 | 0.292 | 0.026 | 0.056 |
| 07 | 0.003 | 0.979 | 0.004 | 0.014 |
| 08 | 0.09 | 0.0 | 0.91 | 0.0 |
| 09 | 0.0 | 0.0 | 0.016 | 0.984 |
| 10 | 0.0 | 0.965 | 0.0 | 0.035 |
| 11 | 0.0 | 0.0 | 0.0 | 1.0 |
| 12 | 0.997 | 0.0 | 0.0 | 0.003 |
| 13 | 0.008 | 0.864 | 0.001 | 0.127 |
| 14 | 0.618 | 0.001 | 0.379 | 0.001 |
| 15 | 0.053 | 0.83 | 0.016 | 0.101 |
| 16 | 0.447 | 0.14 | 0.286 | 0.127 |
| 17 | 0.408 | 0.142 | 0.227 | 0.223 |
| 18 | 0.359 | 0.175 | 0.14 | 0.327 |
| 19 | 0.206 | 0.47 | 0.089 | 0.235 |
| 20 | 0.309 | 0.289 | 0.163 | 0.239 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.613 | 0.304 | -1.295 | 0.604 |
| 02 | 0.799 | -0.956 | 0.1 | -1.249 |
| 03 | -0.708 | -1.214 | -1.782 | 1.113 |
| 04 | 1.188 | -1.131 | -1.614 | -1.614 |
| 05 | 0.676 | -1.286 | 0.41 | -1.384 |
| 06 | 0.913 | 0.155 | -2.193 | -1.47 |
| 07 | -3.971 | 1.359 | -3.729 | -2.73 |
| 08 | -1.011 | -4.761 | 1.286 | -4.761 |
| 09 | -4.761 | -4.761 | -2.654 | 1.364 |
| 10 | -4.761 | 1.344 | -4.761 | -1.913 |
| 11 | -4.761 | -4.761 | -4.761 | 1.38 |
| 12 | 1.377 | -4.761 | -4.761 | -3.971 |
| 13 | -3.232 | 1.234 | -4.29 | -0.667 |
| 14 | 0.9 | -4.29 | 0.414 | -4.29 |
| 15 | -1.515 | 1.194 | -2.654 | -0.891 |
| 16 | 0.577 | -0.571 | 0.132 | -0.667 |
| 17 | 0.486 | -0.562 | -0.094 | -0.112 |
| 18 | 0.36 | -0.355 | -0.576 | 0.265 |
| 19 | -0.191 | 0.627 | -1.014 | -0.062 |
| 20 | 0.209 | 0.145 | -0.421 | -0.046 |
| P-value | Threshold |
|---|---|
| 0.001 | 0.33046 |
| 0.0005 | 1.84096 |
| 0.0001 | 5.00011 |