| Motif | ZNF85.H12RSNP.1.P.D |
| Gene (human) | ZNF85 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 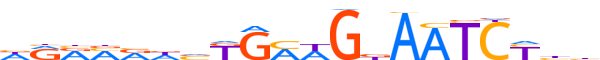 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | D |
| Motif | ZNF85.H12RSNP.1.P.D |
| Gene (human) | ZNF85 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | D |
| Motif length | 20 |
| Consensus | dRRMRdvKGMWGdAATCYbh |
| GC content | 40.83% |
| Information content (bits; total / per base) | 14.564 / 0.728 |
| Data sources | ChIP-Seq |
| Aligned words | 1056 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 2 (9) | 0.869 | 0.971 | 0.823 | 0.969 | 0.897 | 0.968 | 6.148 | 9.054 | 251.569 | 413.032 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | Unclassified {2.3.3.0} (TFClass) |
| TFClass ID | TFClass: 2.3.3.0.198 |
| HGNC | HGNC:13160 |
| MGI | |
| EntrezGene (human) | GeneID:7639 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZNF85_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q03923 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 0 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | ZNF85.H12RSNP.1.P.D.pcm |
| PWM | ZNF85.H12RSNP.1.P.D.pwm |
| PFM | ZNF85.H12RSNP.1.P.D.pfm |
| Alignment | ZNF85.H12RSNP.1.P.D.words.tsv |
| Threshold to P-value map | ZNF85.H12RSNP.1.P.D.thr |
| Motif in other formats | |
| JASPAR format | ZNF85.H12RSNP.1.P.D_jaspar_format.txt |
| MEME format | ZNF85.H12RSNP.1.P.D_meme_format.meme |
| Transfac format | ZNF85.H12RSNP.1.P.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 599.0 | 108.0 | 122.0 | 227.0 |
| 02 | 200.0 | 91.0 | 690.0 | 75.0 |
| 03 | 656.0 | 116.0 | 201.0 | 83.0 |
| 04 | 648.0 | 196.0 | 132.0 | 80.0 |
| 05 | 574.0 | 89.0 | 313.0 | 80.0 |
| 06 | 543.0 | 80.0 | 132.0 | 301.0 |
| 07 | 197.0 | 541.0 | 200.0 | 118.0 |
| 08 | 126.0 | 39.0 | 155.0 | 736.0 |
| 09 | 190.0 | 9.0 | 846.0 | 11.0 |
| 10 | 571.0 | 411.0 | 42.0 | 32.0 |
| 11 | 537.0 | 37.0 | 51.0 | 431.0 |
| 12 | 25.0 | 7.0 | 1015.0 | 9.0 |
| 13 | 130.0 | 94.0 | 377.0 | 455.0 |
| 14 | 1005.0 | 22.0 | 19.0 | 10.0 |
| 15 | 893.0 | 132.0 | 15.0 | 16.0 |
| 16 | 17.0 | 55.0 | 36.0 | 948.0 |
| 17 | 35.0 | 956.0 | 26.0 | 39.0 |
| 18 | 157.0 | 182.0 | 17.0 | 700.0 |
| 19 | 132.0 | 400.0 | 162.0 | 362.0 |
| 20 | 185.0 | 380.0 | 127.0 | 364.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.567 | 0.102 | 0.116 | 0.215 |
| 02 | 0.189 | 0.086 | 0.653 | 0.071 |
| 03 | 0.621 | 0.11 | 0.19 | 0.079 |
| 04 | 0.614 | 0.186 | 0.125 | 0.076 |
| 05 | 0.544 | 0.084 | 0.296 | 0.076 |
| 06 | 0.514 | 0.076 | 0.125 | 0.285 |
| 07 | 0.187 | 0.512 | 0.189 | 0.112 |
| 08 | 0.119 | 0.037 | 0.147 | 0.697 |
| 09 | 0.18 | 0.009 | 0.801 | 0.01 |
| 10 | 0.541 | 0.389 | 0.04 | 0.03 |
| 11 | 0.509 | 0.035 | 0.048 | 0.408 |
| 12 | 0.024 | 0.007 | 0.961 | 0.009 |
| 13 | 0.123 | 0.089 | 0.357 | 0.431 |
| 14 | 0.952 | 0.021 | 0.018 | 0.009 |
| 15 | 0.846 | 0.125 | 0.014 | 0.015 |
| 16 | 0.016 | 0.052 | 0.034 | 0.898 |
| 17 | 0.033 | 0.905 | 0.025 | 0.037 |
| 18 | 0.149 | 0.172 | 0.016 | 0.663 |
| 19 | 0.125 | 0.379 | 0.153 | 0.343 |
| 20 | 0.175 | 0.36 | 0.12 | 0.345 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.816 | -0.884 | -0.764 | -0.15 |
| 02 | -0.276 | -1.053 | 0.957 | -1.242 |
| 03 | 0.906 | -0.814 | -0.271 | -1.143 |
| 04 | 0.894 | -0.296 | -0.687 | -1.179 |
| 05 | 0.773 | -1.075 | 0.169 | -1.179 |
| 06 | 0.718 | -1.179 | -0.687 | 0.13 |
| 07 | -0.291 | 0.714 | -0.276 | -0.797 |
| 08 | -0.733 | -1.875 | -0.528 | 1.021 |
| 09 | -0.326 | -3.208 | 1.16 | -3.038 |
| 10 | 0.768 | 0.44 | -1.804 | -2.064 |
| 11 | 0.707 | -1.926 | -1.617 | 0.488 |
| 12 | -2.296 | -3.415 | 1.342 | -3.208 |
| 13 | -0.702 | -1.021 | 0.354 | 0.542 |
| 14 | 1.332 | -2.415 | -2.55 | -3.119 |
| 15 | 1.214 | -0.687 | -2.765 | -2.707 |
| 16 | -2.652 | -1.544 | -1.952 | 1.274 |
| 17 | -1.979 | 1.282 | -2.26 | -1.875 |
| 18 | -0.515 | -0.369 | -2.652 | 0.971 |
| 19 | -0.687 | 0.413 | -0.484 | 0.314 |
| 20 | -0.353 | 0.362 | -0.725 | 0.319 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.43196 |
| 0.0005 | 4.48236 |
| 0.0001 | 6.69296 |