| Motif | ZN692.H12INVIVO.0.P.C |
| Gene (human) | ZNF692 (GeneCards) |
| Gene synonyms (human) | AREBP, ZFP692 |
| Gene (mouse) | Znf692 |
| Gene synonyms (mouse) | AREBP, Zfp692 |
| LOGO | 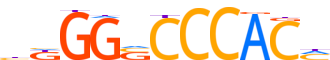 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | C |
| Motif | ZN692.H12INVIVO.0.P.C |
| Gene (human) | ZNF692 (GeneCards) |
| Gene synonyms (human) | AREBP, ZFP692 |
| Gene (mouse) | Znf692 |
| Gene synonyms (mouse) | AREBP, Zfp692 |
| LOGO | 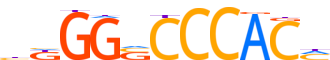 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | C |
| Motif length | 11 |
| Consensus | bKGGSCCCACh |
| GC content | 77.06% |
| Information content (bits; total / per base) | 12.773 / 1.161 |
| Data sources | ChIP-Seq |
| Aligned words | 999 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (7) | 0.823 | 0.854 | 0.782 | 0.814 | 0.773 | 0.806 | 2.87 | 3.096 | 161.721 | 286.284 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | ZFP91-like {2.3.3.3} (TFClass) |
| TFClass ID | TFClass: 2.3.3.3.2 |
| HGNC | HGNC:26049 |
| MGI | MGI:2144276 |
| EntrezGene (human) | GeneID:55657 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:103836 (SSTAR profile) |
| UniProt ID (human) | ZN692_HUMAN |
| UniProt ID (mouse) | ZN692_MOUSE |
| UniProt AC (human) | Q9BU19 (TFClass) |
| UniProt AC (mouse) | Q3U381 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | ZN692.H12INVIVO.0.P.C.pcm |
| PWM | ZN692.H12INVIVO.0.P.C.pwm |
| PFM | ZN692.H12INVIVO.0.P.C.pfm |
| Alignment | ZN692.H12INVIVO.0.P.C.words.tsv |
| Threshold to P-value map | ZN692.H12INVIVO.0.P.C.thr |
| Motif in other formats | |
| JASPAR format | ZN692.H12INVIVO.0.P.C_jaspar_format.txt |
| MEME format | ZN692.H12INVIVO.0.P.C_meme_format.meme |
| Transfac format | ZN692.H12INVIVO.0.P.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 121.0 | 408.0 | 183.0 | 287.0 |
| 02 | 112.0 | 96.0 | 606.0 | 185.0 |
| 03 | 5.0 | 7.0 | 954.0 | 33.0 |
| 04 | 38.0 | 11.0 | 925.0 | 25.0 |
| 05 | 116.0 | 290.0 | 551.0 | 42.0 |
| 06 | 7.0 | 913.0 | 7.0 | 72.0 |
| 07 | 7.0 | 970.0 | 4.0 | 18.0 |
| 08 | 17.0 | 975.0 | 2.0 | 5.0 |
| 09 | 945.0 | 5.0 | 33.0 | 16.0 |
| 10 | 3.0 | 904.0 | 66.0 | 26.0 |
| 11 | 250.0 | 522.0 | 36.0 | 191.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.121 | 0.408 | 0.183 | 0.287 |
| 02 | 0.112 | 0.096 | 0.607 | 0.185 |
| 03 | 0.005 | 0.007 | 0.955 | 0.033 |
| 04 | 0.038 | 0.011 | 0.926 | 0.025 |
| 05 | 0.116 | 0.29 | 0.552 | 0.042 |
| 06 | 0.007 | 0.914 | 0.007 | 0.072 |
| 07 | 0.007 | 0.971 | 0.004 | 0.018 |
| 08 | 0.017 | 0.976 | 0.002 | 0.005 |
| 09 | 0.946 | 0.005 | 0.033 | 0.016 |
| 10 | 0.003 | 0.905 | 0.066 | 0.026 |
| 11 | 0.25 | 0.523 | 0.036 | 0.191 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.717 | 0.488 | -0.308 | 0.138 |
| 02 | -0.794 | -0.945 | 0.882 | -0.298 |
| 03 | -3.621 | -3.361 | 1.335 | -1.98 |
| 04 | -1.845 | -2.984 | 1.304 | -2.242 |
| 05 | -0.759 | 0.148 | 0.788 | -1.749 |
| 06 | -3.361 | 1.291 | -3.361 | -1.227 |
| 07 | -3.361 | 1.352 | -3.782 | -2.545 |
| 08 | -2.597 | 1.357 | -4.212 | -3.621 |
| 09 | 1.326 | -3.621 | -1.98 | -2.652 |
| 10 | -3.974 | 1.281 | -1.312 | -2.205 |
| 11 | 0.001 | 0.734 | -1.897 | -0.266 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.77468 |
| 0.0005 | 4.881275 |
| 0.0001 | 7.034765 |