| Motif | ZN664.H12INVIVO.0.P.C |
| Gene (human) | ZNF664 (GeneCards) |
| Gene synonyms (human) | ZFOC1, ZNF176 |
| Gene (mouse) | Znf664 |
| Gene synonyms (mouse) | Zfp664 |
| LOGO | 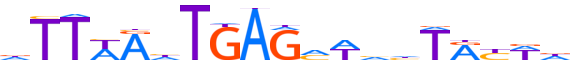 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | C |
| Motif | ZN664.H12INVIVO.0.P.C |
| Gene (human) | ZNF664 (GeneCards) |
| Gene synonyms (human) | ZFOC1, ZNF176 |
| Gene (mouse) | Znf664 |
| Gene synonyms (mouse) | Zfp664 |
| LOGO | 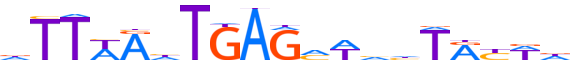 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | C |
| Motif length | 19 |
| Consensus | dTTWAhTGAGhWvhTRYWd |
| GC content | 28.12% |
| Information content (bits; total / per base) | 15.894 / 0.837 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (7) | 0.945 | 0.952 | 0.913 | 0.923 | 0.948 | 0.951 | 6.197 | 6.634 | 396.569 | 459.137 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | Unclassified {2.3.3.0} (TFClass) |
| TFClass ID | TFClass: 2.3.3.0.84 |
| HGNC | HGNC:25406 |
| MGI | MGI:2442505 |
| EntrezGene (human) | GeneID:144348 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:269704 (SSTAR profile) |
| UniProt ID (human) | ZN664_HUMAN |
| UniProt ID (mouse) | ZN664_MOUSE |
| UniProt AC (human) | Q8N3J9 (TFClass) |
| UniProt AC (mouse) | Q4VA44 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | ZN664.H12INVIVO.0.P.C.pcm |
| PWM | ZN664.H12INVIVO.0.P.C.pwm |
| PFM | ZN664.H12INVIVO.0.P.C.pfm |
| Alignment | ZN664.H12INVIVO.0.P.C.words.tsv |
| Threshold to P-value map | ZN664.H12INVIVO.0.P.C.thr |
| Motif in other formats | |
| JASPAR format | ZN664.H12INVIVO.0.P.C_jaspar_format.txt |
| MEME format | ZN664.H12INVIVO.0.P.C_meme_format.meme |
| Transfac format | ZN664.H12INVIVO.0.P.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 467.0 | 102.0 | 206.0 | 225.0 |
| 02 | 13.0 | 74.0 | 2.0 | 911.0 |
| 03 | 62.0 | 3.0 | 13.0 | 922.0 |
| 04 | 527.0 | 47.0 | 8.0 | 418.0 |
| 05 | 818.0 | 46.0 | 94.0 | 42.0 |
| 06 | 436.0 | 159.0 | 82.0 | 323.0 |
| 07 | 6.0 | 7.0 | 2.0 | 985.0 |
| 08 | 34.0 | 12.0 | 868.0 | 86.0 |
| 09 | 969.0 | 6.0 | 6.0 | 19.0 |
| 10 | 91.0 | 11.0 | 847.0 | 51.0 |
| 11 | 404.0 | 439.0 | 69.0 | 88.0 |
| 12 | 434.0 | 18.0 | 31.0 | 517.0 |
| 13 | 480.0 | 221.0 | 157.0 | 142.0 |
| 14 | 208.0 | 458.0 | 123.0 | 211.0 |
| 15 | 98.0 | 25.0 | 30.0 | 847.0 |
| 16 | 695.0 | 34.0 | 160.0 | 111.0 |
| 17 | 74.0 | 529.0 | 76.0 | 321.0 |
| 18 | 224.0 | 57.0 | 96.0 | 623.0 |
| 19 | 473.0 | 86.0 | 139.0 | 302.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.467 | 0.102 | 0.206 | 0.225 |
| 02 | 0.013 | 0.074 | 0.002 | 0.911 |
| 03 | 0.062 | 0.003 | 0.013 | 0.922 |
| 04 | 0.527 | 0.047 | 0.008 | 0.418 |
| 05 | 0.818 | 0.046 | 0.094 | 0.042 |
| 06 | 0.436 | 0.159 | 0.082 | 0.323 |
| 07 | 0.006 | 0.007 | 0.002 | 0.985 |
| 08 | 0.034 | 0.012 | 0.868 | 0.086 |
| 09 | 0.969 | 0.006 | 0.006 | 0.019 |
| 10 | 0.091 | 0.011 | 0.847 | 0.051 |
| 11 | 0.404 | 0.439 | 0.069 | 0.088 |
| 12 | 0.434 | 0.018 | 0.031 | 0.517 |
| 13 | 0.48 | 0.221 | 0.157 | 0.142 |
| 14 | 0.208 | 0.458 | 0.123 | 0.211 |
| 15 | 0.098 | 0.025 | 0.03 | 0.847 |
| 16 | 0.695 | 0.034 | 0.16 | 0.111 |
| 17 | 0.074 | 0.529 | 0.076 | 0.321 |
| 18 | 0.224 | 0.057 | 0.096 | 0.623 |
| 19 | 0.473 | 0.086 | 0.139 | 0.302 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.622 | -0.887 | -0.192 | -0.105 |
| 02 | -2.839 | -1.201 | -4.213 | 1.288 |
| 03 | -1.374 | -3.975 | -2.839 | 1.3 |
| 04 | 0.742 | -1.642 | -3.253 | 0.511 |
| 05 | 1.181 | -1.663 | -0.967 | -1.75 |
| 06 | 0.553 | -0.449 | -1.101 | 0.255 |
| 07 | -3.484 | -3.362 | -4.213 | 1.366 |
| 08 | -1.952 | -2.909 | 1.24 | -1.054 |
| 09 | 1.35 | -3.484 | -3.484 | -2.497 |
| 10 | -0.999 | -2.985 | 1.215 | -1.563 |
| 11 | 0.477 | 0.56 | -1.27 | -1.032 |
| 12 | 0.549 | -2.546 | -2.04 | 0.723 |
| 13 | 0.649 | -0.122 | -0.461 | -0.56 |
| 14 | -0.183 | 0.602 | -0.702 | -0.168 |
| 15 | -0.926 | -2.243 | -2.071 | 1.215 |
| 16 | 1.018 | -1.952 | -0.442 | -0.803 |
| 17 | -1.201 | 0.746 | -1.175 | 0.248 |
| 18 | -0.109 | -1.455 | -0.946 | 0.909 |
| 19 | 0.634 | -1.054 | -0.582 | 0.188 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.77061 |
| 0.0005 | 3.93306 |
| 0.0001 | 6.37206 |