| Motif | ZN567.H12CORE.0.P.C |
| Gene (human) | ZNF567 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 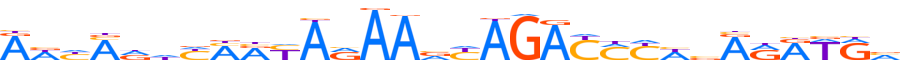 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | C |
| Motif | ZN567.H12CORE.0.P.C |
| Gene (human) | ZNF567 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 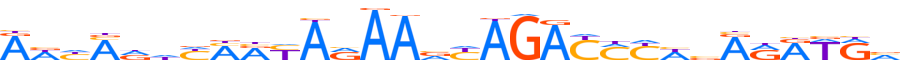 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | C |
| Motif length | 30 |
| Consensus | AWhMddYMRYARAAvYAGAYMYWvRRRWRv |
| GC content | 39.77% |
| Information content (bits; total / per base) | 22.978 / 0.766 |
| Data sources | ChIP-Seq |
| Aligned words | 1002 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (4) | 0.947 | 0.953 | 0.935 | 0.94 | 0.963 | 0.972 | 11.094 | 11.186 | 503.364 | 539.149 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | ZNF25-like {2.3.3.69} (TFClass) |
| TFClass ID | TFClass: 2.3.3.69.4 |
| HGNC | HGNC:28696 |
| MGI | |
| EntrezGene (human) | GeneID:163081 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN567_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q8N184 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | ZN567.H12CORE.0.P.C.pcm |
| PWM | ZN567.H12CORE.0.P.C.pwm |
| PFM | ZN567.H12CORE.0.P.C.pfm |
| Alignment | ZN567.H12CORE.0.P.C.words.tsv |
| Threshold to P-value map | ZN567.H12CORE.0.P.C.thr |
| Motif in other formats | |
| JASPAR format | ZN567.H12CORE.0.P.C_jaspar_format.txt |
| MEME format | ZN567.H12CORE.0.P.C_meme_format.meme |
| Transfac format | ZN567.H12CORE.0.P.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 809.0 | 43.0 | 91.0 | 59.0 |
| 02 | 612.0 | 91.0 | 146.0 | 153.0 |
| 03 | 161.0 | 568.0 | 94.0 | 179.0 |
| 04 | 774.0 | 96.0 | 74.0 | 58.0 |
| 05 | 338.0 | 74.0 | 494.0 | 96.0 |
| 06 | 262.0 | 121.0 | 134.0 | 485.0 |
| 07 | 147.0 | 632.0 | 71.0 | 152.0 |
| 08 | 708.0 | 129.0 | 70.0 | 95.0 |
| 09 | 683.0 | 103.0 | 119.0 | 97.0 |
| 10 | 89.0 | 275.0 | 41.0 | 597.0 |
| 11 | 908.0 | 26.0 | 26.0 | 42.0 |
| 12 | 277.0 | 86.0 | 579.0 | 60.0 |
| 13 | 961.0 | 2.0 | 34.0 | 5.0 |
| 14 | 940.0 | 13.0 | 28.0 | 21.0 |
| 15 | 436.0 | 129.0 | 384.0 | 53.0 |
| 16 | 125.0 | 575.0 | 39.0 | 263.0 |
| 17 | 907.0 | 9.0 | 31.0 | 55.0 |
| 18 | 38.0 | 12.0 | 939.0 | 13.0 |
| 19 | 861.0 | 4.0 | 133.0 | 4.0 |
| 20 | 59.0 | 761.0 | 32.0 | 150.0 |
| 21 | 157.0 | 680.0 | 32.0 | 133.0 |
| 22 | 122.0 | 686.0 | 26.0 | 168.0 |
| 23 | 587.0 | 115.0 | 63.0 | 237.0 |
| 24 | 158.0 | 305.0 | 419.0 | 120.0 |
| 25 | 792.0 | 56.0 | 115.0 | 39.0 |
| 26 | 292.0 | 30.0 | 593.0 | 87.0 |
| 27 | 724.0 | 49.0 | 178.0 | 51.0 |
| 28 | 105.0 | 50.0 | 96.0 | 751.0 |
| 29 | 151.0 | 43.0 | 732.0 | 76.0 |
| 30 | 529.0 | 247.0 | 131.0 | 95.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.807 | 0.043 | 0.091 | 0.059 |
| 02 | 0.611 | 0.091 | 0.146 | 0.153 |
| 03 | 0.161 | 0.567 | 0.094 | 0.179 |
| 04 | 0.772 | 0.096 | 0.074 | 0.058 |
| 05 | 0.337 | 0.074 | 0.493 | 0.096 |
| 06 | 0.261 | 0.121 | 0.134 | 0.484 |
| 07 | 0.147 | 0.631 | 0.071 | 0.152 |
| 08 | 0.707 | 0.129 | 0.07 | 0.095 |
| 09 | 0.682 | 0.103 | 0.119 | 0.097 |
| 10 | 0.089 | 0.274 | 0.041 | 0.596 |
| 11 | 0.906 | 0.026 | 0.026 | 0.042 |
| 12 | 0.276 | 0.086 | 0.578 | 0.06 |
| 13 | 0.959 | 0.002 | 0.034 | 0.005 |
| 14 | 0.938 | 0.013 | 0.028 | 0.021 |
| 15 | 0.435 | 0.129 | 0.383 | 0.053 |
| 16 | 0.125 | 0.574 | 0.039 | 0.262 |
| 17 | 0.905 | 0.009 | 0.031 | 0.055 |
| 18 | 0.038 | 0.012 | 0.937 | 0.013 |
| 19 | 0.859 | 0.004 | 0.133 | 0.004 |
| 20 | 0.059 | 0.759 | 0.032 | 0.15 |
| 21 | 0.157 | 0.679 | 0.032 | 0.133 |
| 22 | 0.122 | 0.685 | 0.026 | 0.168 |
| 23 | 0.586 | 0.115 | 0.063 | 0.237 |
| 24 | 0.158 | 0.304 | 0.418 | 0.12 |
| 25 | 0.79 | 0.056 | 0.115 | 0.039 |
| 26 | 0.291 | 0.03 | 0.592 | 0.087 |
| 27 | 0.723 | 0.049 | 0.178 | 0.051 |
| 28 | 0.105 | 0.05 | 0.096 | 0.75 |
| 29 | 0.151 | 0.043 | 0.731 | 0.076 |
| 30 | 0.528 | 0.247 | 0.131 | 0.095 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1.168 | -1.73 | -1.001 | -1.424 |
| 02 | 0.889 | -1.001 | -0.535 | -0.489 |
| 03 | -0.438 | 0.815 | -0.969 | -0.333 |
| 04 | 1.123 | -0.948 | -1.203 | -1.441 |
| 05 | 0.298 | -1.203 | 0.676 | -0.948 |
| 06 | 0.045 | -0.72 | -0.62 | 0.657 |
| 07 | -0.528 | 0.921 | -1.244 | -0.495 |
| 08 | 1.035 | -0.657 | -1.257 | -0.958 |
| 09 | 0.999 | -0.879 | -0.737 | -0.938 |
| 10 | -1.022 | 0.093 | -1.775 | 0.864 |
| 11 | 1.283 | -2.208 | -2.208 | -1.752 |
| 12 | 0.1 | -1.056 | 0.834 | -1.408 |
| 13 | 1.339 | -4.215 | -1.954 | -3.624 |
| 14 | 1.317 | -2.841 | -2.138 | -2.407 |
| 15 | 0.551 | -0.657 | 0.425 | -1.528 |
| 16 | -0.688 | 0.827 | -1.823 | 0.048 |
| 17 | 1.282 | -3.158 | -2.042 | -1.492 |
| 18 | -1.848 | -2.911 | 1.316 | -2.841 |
| 19 | 1.23 | -3.785 | -0.627 | -3.785 |
| 20 | -1.424 | 1.107 | -2.012 | -0.508 |
| 21 | -0.463 | 0.994 | -2.012 | -0.627 |
| 22 | -0.712 | 1.003 | -2.208 | -0.396 |
| 23 | 0.848 | -0.77 | -1.36 | -0.055 |
| 24 | -0.457 | 0.196 | 0.512 | -0.729 |
| 25 | 1.146 | -1.475 | -0.77 | -1.823 |
| 26 | 0.152 | -2.073 | 0.858 | -1.045 |
| 27 | 1.057 | -1.604 | -0.339 | -1.565 |
| 28 | -0.86 | -1.584 | -0.948 | 1.093 |
| 29 | -0.502 | -1.73 | 1.068 | -1.177 |
| 30 | 0.744 | -0.014 | -0.642 | -0.958 |
| P-value | Threshold |
|---|---|
| 0.001 | 0.31881 |
| 0.0005 | 1.62841 |
| 0.0001 | 4.43086 |