| Motif | ZN557.H12INVITRO.0.P.D |
| Gene (human) | ZNF557 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 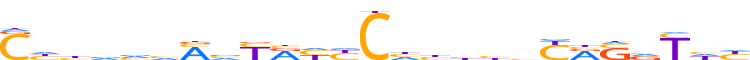 |
| Motif subtype | 0 |
| Quality | D |
| Motif | ZN557.H12INVITRO.0.P.D |
| Gene (human) | ZNF557 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 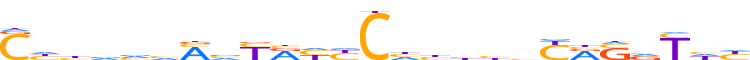 |
| Motif subtype | 0 |
| Quality | D |
| Motif length | 25 |
| Consensus | ddRbSYRvvdvdGvdYRbKdbbddG |
| GC content | 52.32% |
| Information content (bits; total / per base) | 10.979 / 0.439 |
| Data sources | ChIP-Seq |
| Aligned words | 675 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (4) | 0.7 | 0.739 | 0.488 | 0.535 | 0.83 | 0.853 | 4.321 | 5.93 | 93.689 | 96.42 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | ZNF558-like {2.3.3.43} (TFClass) |
| TFClass ID | TFClass: 2.3.3.43.2 |
| HGNC | HGNC:28632 |
| MGI | |
| EntrezGene (human) | GeneID:79230 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN557_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q8N988 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | ZN557.H12INVITRO.0.P.D.pcm |
| PWM | ZN557.H12INVITRO.0.P.D.pwm |
| PFM | ZN557.H12INVITRO.0.P.D.pfm |
| Alignment | ZN557.H12INVITRO.0.P.D.words.tsv |
| Threshold to P-value map | ZN557.H12INVITRO.0.P.D.thr |
| Motif in other formats | |
| JASPAR format | ZN557.H12INVITRO.0.P.D_jaspar_format.txt |
| MEME format | ZN557.H12INVITRO.0.P.D_meme_format.meme |
| Transfac format | ZN557.H12INVITRO.0.P.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 188.0 | 46.0 | 353.0 | 88.0 |
| 02 | 322.0 | 39.0 | 152.0 | 162.0 |
| 03 | 521.0 | 22.0 | 108.0 | 24.0 |
| 04 | 48.0 | 329.0 | 121.0 | 177.0 |
| 05 | 40.0 | 445.0 | 134.0 | 56.0 |
| 06 | 72.0 | 100.0 | 75.0 | 428.0 |
| 07 | 120.0 | 41.0 | 470.0 | 44.0 |
| 08 | 142.0 | 175.0 | 272.0 | 86.0 |
| 09 | 266.0 | 100.0 | 235.0 | 74.0 |
| 10 | 304.0 | 83.0 | 150.0 | 138.0 |
| 11 | 189.0 | 88.0 | 323.0 | 75.0 |
| 12 | 182.0 | 20.0 | 203.0 | 270.0 |
| 13 | 25.0 | 9.0 | 639.0 | 2.0 |
| 14 | 140.0 | 78.0 | 397.0 | 60.0 |
| 15 | 293.0 | 18.0 | 266.0 | 98.0 |
| 16 | 51.0 | 121.0 | 80.0 | 423.0 |
| 17 | 452.0 | 46.0 | 144.0 | 33.0 |
| 18 | 46.0 | 160.0 | 210.0 | 259.0 |
| 19 | 40.0 | 70.0 | 109.0 | 456.0 |
| 20 | 126.0 | 63.0 | 240.0 | 246.0 |
| 21 | 51.0 | 161.0 | 276.0 | 187.0 |
| 22 | 81.0 | 201.0 | 122.0 | 271.0 |
| 23 | 235.0 | 80.0 | 255.0 | 105.0 |
| 24 | 143.0 | 50.0 | 341.0 | 141.0 |
| 25 | 22.0 | 58.0 | 551.0 | 44.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.279 | 0.068 | 0.523 | 0.13 |
| 02 | 0.477 | 0.058 | 0.225 | 0.24 |
| 03 | 0.772 | 0.033 | 0.16 | 0.036 |
| 04 | 0.071 | 0.487 | 0.179 | 0.262 |
| 05 | 0.059 | 0.659 | 0.199 | 0.083 |
| 06 | 0.107 | 0.148 | 0.111 | 0.634 |
| 07 | 0.178 | 0.061 | 0.696 | 0.065 |
| 08 | 0.21 | 0.259 | 0.403 | 0.127 |
| 09 | 0.394 | 0.148 | 0.348 | 0.11 |
| 10 | 0.45 | 0.123 | 0.222 | 0.204 |
| 11 | 0.28 | 0.13 | 0.479 | 0.111 |
| 12 | 0.27 | 0.03 | 0.301 | 0.4 |
| 13 | 0.037 | 0.013 | 0.947 | 0.003 |
| 14 | 0.207 | 0.116 | 0.588 | 0.089 |
| 15 | 0.434 | 0.027 | 0.394 | 0.145 |
| 16 | 0.076 | 0.179 | 0.119 | 0.627 |
| 17 | 0.67 | 0.068 | 0.213 | 0.049 |
| 18 | 0.068 | 0.237 | 0.311 | 0.384 |
| 19 | 0.059 | 0.104 | 0.161 | 0.676 |
| 20 | 0.187 | 0.093 | 0.356 | 0.364 |
| 21 | 0.076 | 0.239 | 0.409 | 0.277 |
| 22 | 0.12 | 0.298 | 0.181 | 0.401 |
| 23 | 0.348 | 0.119 | 0.378 | 0.156 |
| 24 | 0.212 | 0.074 | 0.505 | 0.209 |
| 25 | 0.033 | 0.086 | 0.816 | 0.065 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.107 | -1.275 | 0.733 | -0.642 |
| 02 | 0.642 | -1.434 | -0.103 | -0.04 |
| 03 | 1.121 | -1.976 | -0.441 | -1.894 |
| 04 | -1.233 | 0.663 | -0.329 | 0.047 |
| 05 | -1.409 | 0.964 | -0.228 | -1.084 |
| 06 | -0.839 | -0.517 | -0.799 | 0.925 |
| 07 | -0.337 | -1.385 | 1.018 | -1.317 |
| 08 | -0.171 | 0.036 | 0.474 | -0.665 |
| 09 | 0.452 | -0.517 | 0.328 | -0.812 |
| 10 | 0.584 | -0.7 | -0.117 | -0.199 |
| 11 | 0.112 | -0.642 | 0.645 | -0.799 |
| 12 | 0.075 | -2.064 | 0.183 | 0.466 |
| 13 | -1.856 | -2.774 | 1.324 | -3.849 |
| 14 | -0.185 | -0.761 | 0.85 | -1.017 |
| 15 | 0.548 | -2.161 | 0.452 | -0.537 |
| 16 | -1.175 | -0.329 | -0.736 | 0.913 |
| 17 | 0.979 | -1.275 | -0.157 | -1.593 |
| 18 | -1.275 | -0.053 | 0.217 | 0.425 |
| 19 | -1.409 | -0.867 | -0.432 | 0.988 |
| 20 | -0.289 | -0.969 | 0.349 | 0.374 |
| 21 | -1.175 | -0.047 | 0.488 | 0.102 |
| 22 | -0.724 | 0.173 | -0.321 | 0.47 |
| 23 | 0.328 | -0.736 | 0.41 | -0.469 |
| 24 | -0.164 | -1.194 | 0.699 | -0.178 |
| 25 | -1.976 | -1.05 | 1.177 | -1.317 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.51716 |
| 0.0005 | 5.33736 |
| 0.0001 | 7.06556 |