| Motif | ZN554.H12CORE.0.P.C |
| Gene (human) | ZNF554 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 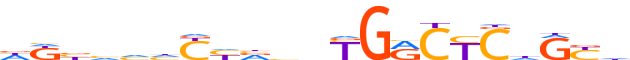 |
| Motif subtype | 0 |
| Quality | C |
| Motif | ZN554.H12CORE.0.P.C |
| Gene (human) | ZNF554 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 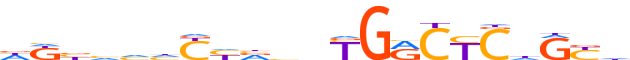 |
| Motif subtype | 0 |
| Quality | C |
| Motif length | 21 |
| Consensus | dRYdGRGYCRnbbdRdddvMd |
| GC content | 55.89% |
| Information content (bits; total / per base) | 11.856 / 0.565 |
| Data sources | ChIP-Seq |
| Aligned words | 992 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (5) | 0.849 | 0.868 | 0.755 | 0.783 | 0.863 | 0.875 | 3.874 | 4.068 | 311.553 | 364.398 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | Unclassified {2.3.3.0} (TFClass) |
| TFClass ID | TFClass: 2.3.3.0.63 |
| HGNC | HGNC:26629 |
| MGI | |
| EntrezGene (human) | GeneID:115196 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN554_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q86TJ5 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | ZN554.H12CORE.0.P.C.pcm |
| PWM | ZN554.H12CORE.0.P.C.pwm |
| PFM | ZN554.H12CORE.0.P.C.pfm |
| Alignment | ZN554.H12CORE.0.P.C.words.tsv |
| Threshold to P-value map | ZN554.H12CORE.0.P.C.thr |
| Motif in other formats | |
| JASPAR format | ZN554.H12CORE.0.P.C_jaspar_format.txt |
| MEME format | ZN554.H12CORE.0.P.C_meme_format.meme |
| Transfac format | ZN554.H12CORE.0.P.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 349.0 | 89.0 | 234.0 | 320.0 |
| 02 | 214.0 | 106.0 | 614.0 | 58.0 |
| 03 | 86.0 | 776.0 | 20.0 | 110.0 |
| 04 | 333.0 | 115.0 | 145.0 | 399.0 |
| 05 | 45.0 | 62.0 | 849.0 | 36.0 |
| 06 | 658.0 | 13.0 | 290.0 | 31.0 |
| 07 | 131.0 | 20.0 | 835.0 | 6.0 |
| 08 | 13.0 | 547.0 | 55.0 | 377.0 |
| 09 | 10.0 | 973.0 | 4.0 | 5.0 |
| 10 | 746.0 | 77.0 | 100.0 | 69.0 |
| 11 | 140.0 | 261.0 | 285.0 | 306.0 |
| 12 | 170.0 | 215.0 | 420.0 | 187.0 |
| 13 | 109.0 | 169.0 | 162.0 | 552.0 |
| 14 | 416.0 | 46.0 | 406.0 | 124.0 |
| 15 | 103.0 | 71.0 | 749.0 | 69.0 |
| 16 | 194.0 | 59.0 | 426.0 | 313.0 |
| 17 | 138.0 | 100.0 | 453.0 | 301.0 |
| 18 | 164.0 | 110.0 | 295.0 | 423.0 |
| 19 | 423.0 | 134.0 | 359.0 | 76.0 |
| 20 | 135.0 | 668.0 | 82.0 | 107.0 |
| 21 | 195.0 | 122.0 | 128.0 | 547.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.352 | 0.09 | 0.236 | 0.323 |
| 02 | 0.216 | 0.107 | 0.619 | 0.058 |
| 03 | 0.087 | 0.782 | 0.02 | 0.111 |
| 04 | 0.336 | 0.116 | 0.146 | 0.402 |
| 05 | 0.045 | 0.063 | 0.856 | 0.036 |
| 06 | 0.663 | 0.013 | 0.292 | 0.031 |
| 07 | 0.132 | 0.02 | 0.842 | 0.006 |
| 08 | 0.013 | 0.551 | 0.055 | 0.38 |
| 09 | 0.01 | 0.981 | 0.004 | 0.005 |
| 10 | 0.752 | 0.078 | 0.101 | 0.07 |
| 11 | 0.141 | 0.263 | 0.287 | 0.308 |
| 12 | 0.171 | 0.217 | 0.423 | 0.189 |
| 13 | 0.11 | 0.17 | 0.163 | 0.556 |
| 14 | 0.419 | 0.046 | 0.409 | 0.125 |
| 15 | 0.104 | 0.072 | 0.755 | 0.07 |
| 16 | 0.196 | 0.059 | 0.429 | 0.316 |
| 17 | 0.139 | 0.101 | 0.457 | 0.303 |
| 18 | 0.165 | 0.111 | 0.297 | 0.426 |
| 19 | 0.426 | 0.135 | 0.362 | 0.077 |
| 20 | 0.136 | 0.673 | 0.083 | 0.108 |
| 21 | 0.197 | 0.123 | 0.129 | 0.551 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.34 | -1.013 | -0.058 | 0.253 |
| 02 | -0.146 | -0.841 | 0.902 | -1.431 |
| 03 | -1.046 | 1.136 | -2.442 | -0.804 |
| 04 | 0.293 | -0.761 | -0.532 | 0.473 |
| 05 | -1.676 | -1.366 | 1.226 | -1.89 |
| 06 | 0.971 | -2.831 | 0.155 | -2.032 |
| 07 | -0.632 | -2.442 | 1.209 | -3.476 |
| 08 | -2.831 | 0.787 | -1.482 | 0.416 |
| 09 | -3.059 | 1.362 | -3.776 | -3.615 |
| 10 | 1.097 | -1.154 | -0.898 | -1.262 |
| 11 | -0.566 | 0.051 | 0.138 | 0.209 |
| 12 | -0.374 | -0.142 | 0.524 | -0.28 |
| 13 | -0.813 | -0.38 | -0.422 | 0.796 |
| 14 | 0.514 | -1.655 | 0.49 | -0.686 |
| 15 | -0.869 | -1.234 | 1.101 | -1.262 |
| 16 | -0.244 | -1.414 | 0.538 | 0.231 |
| 17 | -0.581 | -0.898 | 0.599 | 0.192 |
| 18 | -0.41 | -0.804 | 0.172 | 0.531 |
| 19 | 0.531 | -0.61 | 0.368 | -1.167 |
| 20 | -0.602 | 0.987 | -1.093 | -0.832 |
| 21 | -0.239 | -0.702 | -0.655 | 0.787 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.31196 |
| 0.0005 | 5.20776 |
| 0.0001 | 7.07541 |