| Motif | ZN530.H12INVIVO.0.P.B |
| Gene (human) | ZNF530 (GeneCards) |
| Gene synonyms (human) | KIAA1508 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 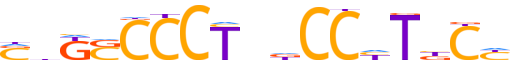 |
| LOGO (reverse complement) | 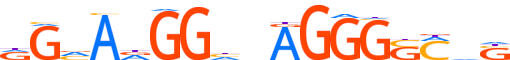 |
| Motif subtype | 0 |
| Quality | B |
| Motif | ZN530.H12INVIVO.0.P.B |
| Gene (human) | ZNF530 (GeneCards) |
| Gene synonyms (human) | KIAA1508 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 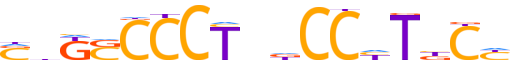 |
| LOGO (reverse complement) | 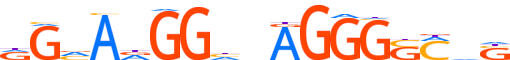 |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 17 |
| Consensus | ShKSCCCTndCChTbCY |
| GC content | 66.85% |
| Information content (bits; total / per base) | 16.029 / 0.943 |
| Data sources | ChIP-Seq |
| Aligned words | 540 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 2 (10) | 0.832 | 0.905 | 0.71 | 0.805 | 0.856 | 0.95 | 5.03 | 5.865 | 57.886 | 246.481 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | Unclassified {2.3.3.0} (TFClass) |
| TFClass ID | TFClass: 2.3.3.0.153 |
| HGNC | HGNC:29297 |
| MGI | |
| EntrezGene (human) | GeneID:348327 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN530_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q6P9A1 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 0 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | ZN530.H12INVIVO.0.P.B.pcm |
| PWM | ZN530.H12INVIVO.0.P.B.pwm |
| PFM | ZN530.H12INVIVO.0.P.B.pfm |
| Alignment | ZN530.H12INVIVO.0.P.B.words.tsv |
| Threshold to P-value map | ZN530.H12INVIVO.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | ZN530.H12INVIVO.0.P.B_jaspar_format.txt |
| MEME format | ZN530.H12INVIVO.0.P.B_meme_format.meme |
| Transfac format | ZN530.H12INVIVO.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 60.0 | 364.0 | 67.0 | 49.0 |
| 02 | 87.0 | 152.0 | 86.0 | 215.0 |
| 03 | 32.0 | 24.0 | 405.0 | 79.0 |
| 04 | 28.0 | 319.0 | 184.0 | 9.0 |
| 05 | 13.0 | 494.0 | 17.0 | 16.0 |
| 06 | 2.0 | 492.0 | 2.0 | 44.0 |
| 07 | 1.0 | 530.0 | 5.0 | 4.0 |
| 08 | 25.0 | 44.0 | 5.0 | 466.0 |
| 09 | 80.0 | 153.0 | 128.0 | 179.0 |
| 10 | 92.0 | 31.0 | 206.0 | 211.0 |
| 11 | 3.0 | 526.0 | 2.0 | 9.0 |
| 12 | 2.0 | 516.0 | 6.0 | 16.0 |
| 13 | 100.0 | 160.0 | 22.0 | 258.0 |
| 14 | 8.0 | 3.0 | 24.0 | 505.0 |
| 15 | 28.0 | 79.0 | 225.0 | 208.0 |
| 16 | 7.0 | 472.0 | 30.0 | 31.0 |
| 17 | 77.0 | 323.0 | 41.0 | 99.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.111 | 0.674 | 0.124 | 0.091 |
| 02 | 0.161 | 0.281 | 0.159 | 0.398 |
| 03 | 0.059 | 0.044 | 0.75 | 0.146 |
| 04 | 0.052 | 0.591 | 0.341 | 0.017 |
| 05 | 0.024 | 0.915 | 0.031 | 0.03 |
| 06 | 0.004 | 0.911 | 0.004 | 0.081 |
| 07 | 0.002 | 0.981 | 0.009 | 0.007 |
| 08 | 0.046 | 0.081 | 0.009 | 0.863 |
| 09 | 0.148 | 0.283 | 0.237 | 0.331 |
| 10 | 0.17 | 0.057 | 0.381 | 0.391 |
| 11 | 0.006 | 0.974 | 0.004 | 0.017 |
| 12 | 0.004 | 0.956 | 0.011 | 0.03 |
| 13 | 0.185 | 0.296 | 0.041 | 0.478 |
| 14 | 0.015 | 0.006 | 0.044 | 0.935 |
| 15 | 0.052 | 0.146 | 0.417 | 0.385 |
| 16 | 0.013 | 0.874 | 0.056 | 0.057 |
| 17 | 0.143 | 0.598 | 0.076 | 0.183 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.797 | 0.985 | -0.689 | -0.993 |
| 02 | -0.433 | 0.117 | -0.444 | 0.461 |
| 03 | -1.403 | -1.675 | 1.091 | -0.528 |
| 04 | -1.53 | 0.853 | 0.307 | -2.559 |
| 05 | -2.238 | 1.289 | -1.995 | -2.051 |
| 06 | -3.643 | 1.285 | -3.643 | -1.098 |
| 07 | -3.972 | 1.359 | -3.034 | -3.199 |
| 08 | -1.637 | -1.098 | -3.034 | 1.231 |
| 09 | -0.515 | 0.124 | -0.053 | 0.279 |
| 10 | -0.378 | -1.433 | 0.419 | 0.442 |
| 11 | -3.397 | 1.351 | -3.643 | -2.559 |
| 12 | -3.643 | 1.332 | -2.892 | -2.051 |
| 13 | -0.296 | 0.168 | -1.757 | 0.642 |
| 14 | -2.658 | -3.397 | -1.675 | 1.311 |
| 15 | -1.53 | -0.528 | 0.506 | 0.428 |
| 16 | -2.768 | 1.243 | -1.465 | -1.433 |
| 17 | -0.553 | 0.866 | -1.166 | -0.306 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.71451 |
| 0.0005 | 3.88621 |
| 0.0001 | 6.35181 |