| Motif | ZN480.H12INVIVO.0.P.C |
| Gene (human) | ZNF480 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 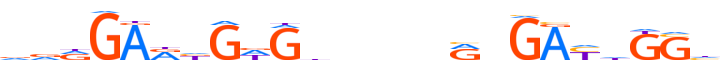 |
| Motif subtype | 0 |
| Quality | C |
| Motif | ZN480.H12INVIVO.0.P.C |
| Gene (human) | ZNF480 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 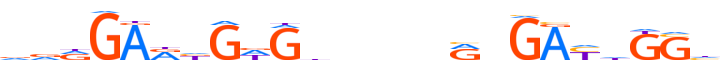 |
| Motif subtype | 0 |
| Quality | C |
| Motif length | 24 |
| Consensus | dSCdRTChYhvnnvCnCvYTChnh |
| GC content | 52.84% |
| Information content (bits; total / per base) | 14.033 / 0.585 |
| Data sources | ChIP-Seq |
| Aligned words | 331 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (4) | 0.958 | 0.965 | 0.949 | 0.955 | 0.961 | 0.967 | 7.25 | 7.293 | 556.952 | 605.886 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | Unclassified {2.3.3.0} (TFClass) |
| TFClass ID | TFClass: 2.3.3.0.134 |
| HGNC | HGNC:23305 |
| MGI | |
| EntrezGene (human) | GeneID:147657 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN480_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q8WV37 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | ZN480.H12INVIVO.0.P.C.pcm |
| PWM | ZN480.H12INVIVO.0.P.C.pwm |
| PFM | ZN480.H12INVIVO.0.P.C.pfm |
| Alignment | ZN480.H12INVIVO.0.P.C.words.tsv |
| Threshold to P-value map | ZN480.H12INVIVO.0.P.C.thr |
| Motif in other formats | |
| JASPAR format | ZN480.H12INVIVO.0.P.C_jaspar_format.txt |
| MEME format | ZN480.H12INVIVO.0.P.C_meme_format.meme |
| Transfac format | ZN480.H12INVIVO.0.P.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 71.0 | 33.0 | 148.0 | 79.0 |
| 02 | 9.0 | 261.0 | 38.0 | 23.0 |
| 03 | 29.0 | 271.0 | 9.0 | 22.0 |
| 04 | 147.0 | 48.0 | 54.0 | 82.0 |
| 05 | 178.0 | 9.0 | 111.0 | 33.0 |
| 06 | 2.0 | 22.0 | 21.0 | 286.0 |
| 07 | 2.0 | 306.0 | 9.0 | 14.0 |
| 08 | 86.0 | 113.0 | 40.0 | 92.0 |
| 09 | 10.0 | 204.0 | 30.0 | 87.0 |
| 10 | 76.0 | 110.0 | 39.0 | 106.0 |
| 11 | 99.0 | 88.0 | 105.0 | 39.0 |
| 12 | 80.0 | 80.0 | 101.0 | 70.0 |
| 13 | 98.0 | 100.0 | 45.0 | 88.0 |
| 14 | 131.0 | 82.0 | 66.0 | 52.0 |
| 15 | 20.0 | 284.0 | 2.0 | 25.0 |
| 16 | 136.0 | 48.0 | 11.0 | 136.0 |
| 17 | 8.0 | 288.0 | 2.0 | 33.0 |
| 18 | 178.0 | 58.0 | 50.0 | 45.0 |
| 19 | 39.0 | 76.0 | 19.0 | 197.0 |
| 20 | 13.0 | 18.0 | 8.0 | 292.0 |
| 21 | 1.0 | 314.0 | 4.0 | 12.0 |
| 22 | 59.0 | 175.0 | 16.0 | 81.0 |
| 23 | 36.0 | 120.0 | 36.0 | 139.0 |
| 24 | 64.0 | 83.0 | 43.0 | 141.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.215 | 0.1 | 0.447 | 0.239 |
| 02 | 0.027 | 0.789 | 0.115 | 0.069 |
| 03 | 0.088 | 0.819 | 0.027 | 0.066 |
| 04 | 0.444 | 0.145 | 0.163 | 0.248 |
| 05 | 0.538 | 0.027 | 0.335 | 0.1 |
| 06 | 0.006 | 0.066 | 0.063 | 0.864 |
| 07 | 0.006 | 0.924 | 0.027 | 0.042 |
| 08 | 0.26 | 0.341 | 0.121 | 0.278 |
| 09 | 0.03 | 0.616 | 0.091 | 0.263 |
| 10 | 0.23 | 0.332 | 0.118 | 0.32 |
| 11 | 0.299 | 0.266 | 0.317 | 0.118 |
| 12 | 0.242 | 0.242 | 0.305 | 0.211 |
| 13 | 0.296 | 0.302 | 0.136 | 0.266 |
| 14 | 0.396 | 0.248 | 0.199 | 0.157 |
| 15 | 0.06 | 0.858 | 0.006 | 0.076 |
| 16 | 0.411 | 0.145 | 0.033 | 0.411 |
| 17 | 0.024 | 0.87 | 0.006 | 0.1 |
| 18 | 0.538 | 0.175 | 0.151 | 0.136 |
| 19 | 0.118 | 0.23 | 0.057 | 0.595 |
| 20 | 0.039 | 0.054 | 0.024 | 0.882 |
| 21 | 0.003 | 0.949 | 0.012 | 0.036 |
| 22 | 0.178 | 0.529 | 0.048 | 0.245 |
| 23 | 0.109 | 0.363 | 0.109 | 0.42 |
| 24 | 0.193 | 0.251 | 0.13 | 0.426 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.15 | -0.894 | 0.574 | -0.046 |
| 02 | -2.087 | 1.137 | -0.758 | -1.237 |
| 03 | -1.017 | 1.174 | -2.087 | -1.278 |
| 04 | 0.567 | -0.532 | -0.418 | -0.009 |
| 05 | 0.757 | -2.087 | 0.289 | -0.894 |
| 06 | -3.195 | -1.278 | -1.322 | 1.228 |
| 07 | -3.195 | 1.295 | -2.087 | -1.696 |
| 08 | 0.038 | 0.307 | -0.709 | 0.104 |
| 09 | -1.995 | 0.892 | -0.985 | 0.049 |
| 10 | -0.084 | 0.28 | -0.733 | 0.244 |
| 11 | 0.176 | 0.06 | 0.234 | -0.733 |
| 12 | -0.033 | -0.033 | 0.196 | -0.164 |
| 13 | 0.166 | 0.186 | -0.595 | 0.06 |
| 14 | 0.453 | -0.009 | -0.222 | -0.454 |
| 15 | -1.367 | 1.221 | -3.195 | -1.158 |
| 16 | 0.49 | -0.532 | -1.911 | 0.49 |
| 17 | -2.187 | 1.235 | -3.195 | -0.894 |
| 18 | 0.757 | -0.348 | -0.493 | -0.595 |
| 19 | -0.733 | -0.084 | -1.415 | 0.857 |
| 20 | -1.762 | -1.465 | -2.187 | 1.249 |
| 21 | -3.537 | 1.321 | -2.737 | -1.834 |
| 22 | -0.331 | 0.74 | -1.574 | -0.021 |
| 23 | -0.81 | 0.366 | -0.81 | 0.512 |
| 24 | -0.252 | 0.003 | -0.639 | 0.526 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.76311 |
| 0.0005 | 4.75256 |
| 0.0001 | 6.83891 |