| Motif | ZN449.H12INVIVO.1.S.C |
| Gene (human) | ZNF449 (GeneCards) |
| Gene synonyms (human) | ZSCAN19 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 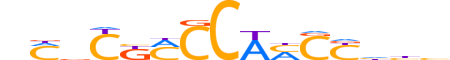 |
| Motif subtype | 1 |
| Quality | C |
| Motif | ZN449.H12INVIVO.1.S.C |
| Gene (human) | ZNF449 (GeneCards) |
| Gene synonyms (human) | ZSCAN19 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 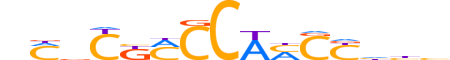 |
| Motif subtype | 1 |
| Quality | C |
| Motif length | 15 |
| Consensus | dddRGKWGGKMGbKn |
| GC content | 64.97% |
| Information content (bits; total / per base) | 10.222 / 0.681 |
| Data sources | HT-SELEX |
| Aligned words | 6585 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 2 (12) | 0.658 | 0.684 | 0.454 | 0.481 | 0.694 | 0.737 | 1.946 | 2.189 | 65.204 | 93.0 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 1 experiments | median | 0.714 | 0.654 | 0.66 | 0.617 | 0.609 | 0.585 |
| best | 0.714 | 0.654 | 0.66 | 0.617 | 0.609 | 0.585 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 2.844 | 0.939 | 0.067 | 0.04 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | Unclassified {2.3.3.0} (TFClass) |
| TFClass ID | TFClass: 2.3.3.0.41 |
| HGNC | HGNC:21039 |
| MGI | |
| EntrezGene (human) | GeneID:203523 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN449_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q6P9G9 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 0 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 0 |
| PCM | ZN449.H12INVIVO.1.S.C.pcm |
| PWM | ZN449.H12INVIVO.1.S.C.pwm |
| PFM | ZN449.H12INVIVO.1.S.C.pfm |
| Alignment | ZN449.H12INVIVO.1.S.C.words.tsv |
| Threshold to P-value map | ZN449.H12INVIVO.1.S.C.thr |
| Motif in other formats | |
| JASPAR format | ZN449.H12INVIVO.1.S.C_jaspar_format.txt |
| MEME format | ZN449.H12INVIVO.1.S.C_meme_format.meme |
| Transfac format | ZN449.H12INVIVO.1.S.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1335.75 | 1040.75 | 2859.75 | 1348.75 |
| 02 | 2090.75 | 616.75 | 2636.75 | 1240.75 |
| 03 | 2306.0 | 568.0 | 2066.0 | 1645.0 |
| 04 | 693.0 | 579.0 | 4735.0 | 578.0 |
| 05 | 199.0 | 705.0 | 5288.0 | 393.0 |
| 06 | 768.0 | 24.0 | 2756.0 | 3037.0 |
| 07 | 2002.0 | 1.0 | 145.0 | 4437.0 |
| 08 | 0.0 | 1.0 | 6584.0 | 0.0 |
| 09 | 2.0 | 950.0 | 5582.0 | 51.0 |
| 10 | 466.0 | 40.0 | 3959.0 | 2120.0 |
| 11 | 1044.0 | 4286.0 | 888.0 | 367.0 |
| 12 | 1067.0 | 0.0 | 5212.0 | 306.0 |
| 13 | 659.0 | 2205.0 | 1844.0 | 1877.0 |
| 14 | 673.25 | 405.25 | 4593.25 | 913.25 |
| 15 | 1614.5 | 1104.5 | 2500.5 | 1365.5 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.203 | 0.158 | 0.434 | 0.205 |
| 02 | 0.318 | 0.094 | 0.4 | 0.188 |
| 03 | 0.35 | 0.086 | 0.314 | 0.25 |
| 04 | 0.105 | 0.088 | 0.719 | 0.088 |
| 05 | 0.03 | 0.107 | 0.803 | 0.06 |
| 06 | 0.117 | 0.004 | 0.419 | 0.461 |
| 07 | 0.304 | 0.0 | 0.022 | 0.674 |
| 08 | 0.0 | 0.0 | 1.0 | 0.0 |
| 09 | 0.0 | 0.144 | 0.848 | 0.008 |
| 10 | 0.071 | 0.006 | 0.601 | 0.322 |
| 11 | 0.159 | 0.651 | 0.135 | 0.056 |
| 12 | 0.162 | 0.0 | 0.791 | 0.046 |
| 13 | 0.1 | 0.335 | 0.28 | 0.285 |
| 14 | 0.102 | 0.062 | 0.698 | 0.139 |
| 15 | 0.245 | 0.168 | 0.38 | 0.207 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.209 | -0.458 | 0.552 | -0.199 |
| 02 | 0.239 | -0.98 | 0.471 | -0.282 |
| 03 | 0.337 | -1.062 | 0.227 | -0.001 |
| 04 | -0.863 | -1.042 | 1.056 | -1.044 |
| 05 | -2.103 | -0.846 | 1.166 | -1.428 |
| 06 | -0.761 | -4.142 | 0.515 | 0.612 |
| 07 | 0.195 | -6.245 | -2.416 | 0.991 |
| 08 | -6.62 | -6.245 | 1.385 | -6.62 |
| 09 | -5.973 | -0.549 | 1.22 | -3.434 |
| 10 | -1.259 | -3.665 | 0.877 | 0.253 |
| 11 | -0.455 | 0.956 | -0.616 | -1.496 |
| 12 | -0.433 | -6.62 | 1.152 | -1.677 |
| 13 | -0.914 | 0.292 | 0.113 | 0.131 |
| 14 | -0.892 | -1.398 | 1.025 | -0.588 |
| 15 | -0.019 | -0.398 | 0.418 | -0.187 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.77031 |
| 0.0005 | 5.61881 |
| 0.0001 | 7.29366 |