| Motif | ZN416.H12RSNP.0.P.D |
| Gene (human) | ZNF416 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 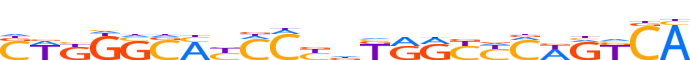 |
| Motif subtype | 0 |
| Quality | D |
| Motif | ZN416.H12RSNP.0.P.D |
| Gene (human) | ZNF416 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 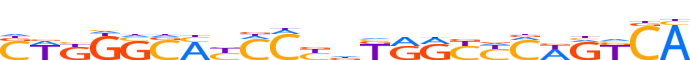 |
| Motif subtype | 0 |
| Quality | D |
| Motif length | 23 |
| Consensus | TGMYWKRRYYMhRGKRKSSCMWS |
| GC content | 60.98% |
| Information content (bits; total / per base) | 17.521 / 0.762 |
| Data sources | ChIP-Seq |
| Aligned words | 78 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (4) | 0.696 | 0.772 | 0.643 | 0.747 | 0.677 | 0.742 | 6.641 | 9.285 | 25.333 | 38.824 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | Unclassified {2.3.3.0} (TFClass) |
| TFClass ID | TFClass: 2.3.3.0.127 |
| HGNC | HGNC:20645 |
| MGI | |
| EntrezGene (human) | GeneID:55659 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN416_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q9BWM5 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | ZN416.H12RSNP.0.P.D.pcm |
| PWM | ZN416.H12RSNP.0.P.D.pwm |
| PFM | ZN416.H12RSNP.0.P.D.pfm |
| Alignment | ZN416.H12RSNP.0.P.D.words.tsv |
| Threshold to P-value map | ZN416.H12RSNP.0.P.D.thr |
| Motif in other formats | |
| JASPAR format | ZN416.H12RSNP.0.P.D_jaspar_format.txt |
| MEME format | ZN416.H12RSNP.0.P.D_meme_format.meme |
| Transfac format | ZN416.H12RSNP.0.P.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 3.0 | 0.0 | 7.0 | 68.0 |
| 02 | 4.0 | 5.0 | 68.0 | 1.0 |
| 03 | 55.0 | 9.0 | 8.0 | 6.0 |
| 04 | 5.0 | 58.0 | 7.0 | 8.0 |
| 05 | 18.0 | 6.0 | 5.0 | 49.0 |
| 06 | 3.0 | 6.0 | 59.0 | 10.0 |
| 07 | 16.0 | 5.0 | 50.0 | 7.0 |
| 08 | 19.0 | 7.0 | 51.0 | 1.0 |
| 09 | 4.0 | 53.0 | 2.0 | 19.0 |
| 10 | 2.0 | 45.0 | 1.0 | 30.0 |
| 11 | 56.0 | 11.0 | 6.0 | 5.0 |
| 12 | 19.0 | 34.0 | 8.0 | 17.0 |
| 13 | 25.0 | 9.0 | 41.0 | 3.0 |
| 14 | 4.0 | 2.0 | 63.0 | 9.0 |
| 15 | 5.0 | 5.0 | 60.0 | 8.0 |
| 16 | 28.0 | 6.0 | 39.0 | 5.0 |
| 17 | 6.0 | 1.0 | 16.0 | 55.0 |
| 18 | 3.0 | 9.0 | 59.0 | 7.0 |
| 19 | 4.0 | 58.0 | 11.0 | 5.0 |
| 20 | 7.0 | 62.0 | 3.0 | 6.0 |
| 21 | 12.0 | 53.0 | 9.0 | 4.0 |
| 22 | 54.0 | 5.0 | 4.0 | 15.0 |
| 23 | 5.0 | 9.0 | 59.0 | 5.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.038 | 0.0 | 0.09 | 0.872 |
| 02 | 0.051 | 0.064 | 0.872 | 0.013 |
| 03 | 0.705 | 0.115 | 0.103 | 0.077 |
| 04 | 0.064 | 0.744 | 0.09 | 0.103 |
| 05 | 0.231 | 0.077 | 0.064 | 0.628 |
| 06 | 0.038 | 0.077 | 0.756 | 0.128 |
| 07 | 0.205 | 0.064 | 0.641 | 0.09 |
| 08 | 0.244 | 0.09 | 0.654 | 0.013 |
| 09 | 0.051 | 0.679 | 0.026 | 0.244 |
| 10 | 0.026 | 0.577 | 0.013 | 0.385 |
| 11 | 0.718 | 0.141 | 0.077 | 0.064 |
| 12 | 0.244 | 0.436 | 0.103 | 0.218 |
| 13 | 0.321 | 0.115 | 0.526 | 0.038 |
| 14 | 0.051 | 0.026 | 0.808 | 0.115 |
| 15 | 0.064 | 0.064 | 0.769 | 0.103 |
| 16 | 0.359 | 0.077 | 0.5 | 0.064 |
| 17 | 0.077 | 0.013 | 0.205 | 0.705 |
| 18 | 0.038 | 0.115 | 0.756 | 0.09 |
| 19 | 0.051 | 0.744 | 0.141 | 0.064 |
| 20 | 0.09 | 0.795 | 0.038 | 0.077 |
| 21 | 0.154 | 0.679 | 0.115 | 0.051 |
| 22 | 0.692 | 0.064 | 0.051 | 0.192 |
| 23 | 0.064 | 0.115 | 0.756 | 0.064 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -1.616 | -2.939 | -0.934 | 1.211 |
| 02 | -1.398 | -1.218 | 1.211 | -2.288 |
| 03 | 1.002 | -0.713 | -0.818 | -1.066 |
| 04 | -1.218 | 1.054 | -0.934 | -0.818 |
| 05 | -0.076 | -1.066 | -1.218 | 0.889 |
| 06 | -1.616 | -1.066 | 1.071 | -0.619 |
| 07 | -0.186 | -1.218 | 0.909 | -0.934 |
| 08 | -0.025 | -0.934 | 0.928 | -2.288 |
| 09 | -1.398 | 0.966 | -1.897 | -0.025 |
| 10 | -1.897 | 0.806 | -2.288 | 0.412 |
| 11 | 1.02 | -0.532 | -1.066 | -1.218 |
| 12 | -0.025 | 0.533 | -0.818 | -0.129 |
| 13 | 0.237 | -0.713 | 0.715 | -1.616 |
| 14 | -1.398 | -1.897 | 1.136 | -0.713 |
| 15 | -1.218 | -1.218 | 1.088 | -0.818 |
| 16 | 0.346 | -1.066 | 0.666 | -1.218 |
| 17 | -1.066 | -2.288 | -0.186 | 1.002 |
| 18 | -1.616 | -0.713 | 1.071 | -0.934 |
| 19 | -1.398 | 1.054 | -0.532 | -1.218 |
| 20 | -0.934 | 1.12 | -1.616 | -1.066 |
| 21 | -0.453 | 0.966 | -0.713 | -1.398 |
| 22 | 0.984 | -1.218 | -1.398 | -0.247 |
| 23 | -1.218 | -0.713 | 1.071 | -1.218 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.32746 |
| 0.0005 | 4.32461 |
| 0.0001 | 6.46101 |