| Motif | ZN333.H12CORE.0.P.C |
| Gene (human) | ZNF333 (GeneCards) |
| Gene synonyms (human) | KIAA1806 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 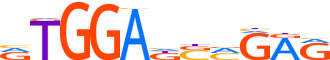 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | C |
| Motif | ZN333.H12CORE.0.P.C |
| Gene (human) | ZNF333 (GeneCards) |
| Gene synonyms (human) | KIAA1806 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 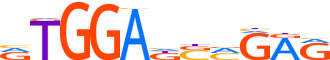 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | C |
| Motif length | 11 |
| Consensus | RTGGAdSMGRR |
| GC content | 61.54% |
| Information content (bits; total / per base) | 11.698 / 1.063 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (4) | 0.862 | 0.874 | 0.77 | 0.788 | 0.862 | 0.866 | 3.648 | 3.712 | 205.257 | 233.013 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | ZNF177-like {2.3.3.40} (TFClass) |
| TFClass ID | TFClass: 2.3.3.40.3 |
| HGNC | HGNC:15624 |
| MGI | |
| EntrezGene (human) | GeneID:84449 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN333_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q96JL9 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | ZN333.H12CORE.0.P.C.pcm |
| PWM | ZN333.H12CORE.0.P.C.pwm |
| PFM | ZN333.H12CORE.0.P.C.pfm |
| Alignment | ZN333.H12CORE.0.P.C.words.tsv |
| Threshold to P-value map | ZN333.H12CORE.0.P.C.thr |
| Motif in other formats | |
| JASPAR format | ZN333.H12CORE.0.P.C_jaspar_format.txt |
| MEME format | ZN333.H12CORE.0.P.C_meme_format.meme |
| Transfac format | ZN333.H12CORE.0.P.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 288.0 | 177.0 | 527.0 | 8.0 |
| 02 | 30.0 | 13.0 | 49.0 | 908.0 |
| 03 | 3.0 | 1.0 | 994.0 | 2.0 |
| 04 | 7.0 | 4.0 | 988.0 | 1.0 |
| 05 | 965.0 | 25.0 | 7.0 | 3.0 |
| 06 | 149.0 | 97.0 | 597.0 | 157.0 |
| 07 | 22.0 | 590.0 | 253.0 | 135.0 |
| 08 | 380.0 | 487.0 | 91.0 | 42.0 |
| 09 | 67.0 | 63.0 | 829.0 | 41.0 |
| 10 | 767.0 | 32.0 | 159.0 | 42.0 |
| 11 | 173.0 | 59.0 | 727.0 | 41.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.288 | 0.177 | 0.527 | 0.008 |
| 02 | 0.03 | 0.013 | 0.049 | 0.908 |
| 03 | 0.003 | 0.001 | 0.994 | 0.002 |
| 04 | 0.007 | 0.004 | 0.988 | 0.001 |
| 05 | 0.965 | 0.025 | 0.007 | 0.003 |
| 06 | 0.149 | 0.097 | 0.597 | 0.157 |
| 07 | 0.022 | 0.59 | 0.253 | 0.135 |
| 08 | 0.38 | 0.487 | 0.091 | 0.042 |
| 09 | 0.067 | 0.063 | 0.829 | 0.041 |
| 10 | 0.767 | 0.032 | 0.159 | 0.042 |
| 11 | 0.173 | 0.059 | 0.727 | 0.041 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.141 | -0.342 | 0.742 | -3.253 |
| 02 | -2.071 | -2.839 | -1.602 | 1.285 |
| 03 | -3.975 | -4.525 | 1.375 | -4.213 |
| 04 | -3.362 | -3.783 | 1.369 | -4.525 |
| 05 | 1.346 | -2.243 | -3.362 | -3.975 |
| 06 | -0.513 | -0.936 | 0.866 | -0.461 |
| 07 | -2.362 | 0.855 | 0.012 | -0.61 |
| 08 | 0.416 | 0.663 | -0.999 | -1.75 |
| 09 | -1.298 | -1.358 | 1.194 | -1.774 |
| 10 | 1.116 | -2.01 | -0.449 | -1.75 |
| 11 | -0.365 | -1.422 | 1.063 | -1.774 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.26245 |
| 0.0005 | 5.254725 |
| 0.0001 | 7.289585 |