| Motif | ZN281.H12INVIVO.1.SM.B |
| Gene (human) | ZNF281 (GeneCards) |
| Gene synonyms (human) | GZP1, ZBP99 |
| Gene (mouse) | Znf281 |
| Gene synonyms (mouse) | Zfp281 |
| LOGO |  |
| LOGO (reverse complement) | 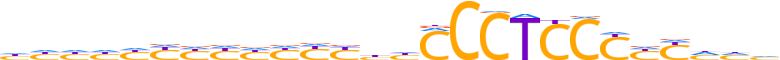 |
| Motif subtype | 1 |
| Quality | B |
| Motif | ZN281.H12INVIVO.1.SM.B |
| Gene (human) | ZNF281 (GeneCards) |
| Gene synonyms (human) | GZP1, ZBP99 |
| Gene (mouse) | Znf281 |
| Gene synonyms (mouse) | Zfp281 |
| LOGO |  |
| LOGO (reverse complement) | 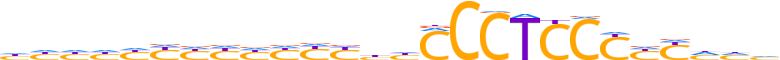 |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 26 |
| Consensus | dddRRRGGAGGGdvRRRRRRRRdddd |
| GC content | 70.22% |
| Information content (bits; total / per base) | 17.958 / 0.691 |
| Data sources | HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 7177 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (5) | 0.737 | 0.751 | 0.557 | 0.585 | 0.807 | 0.827 | 3.338 | 3.51 | 43.602 | 60.244 |
| Mouse | 6 (42) | 0.874 | 0.905 | 0.737 | 0.796 | 0.929 | 0.945 | 4.731 | 5.31 | 140.969 | 221.959 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.706 | 0.644 | 0.609 | 0.591 | 0.538 | 0.55 |
| best | 0.714 | 0.652 | 0.616 | 0.597 | 0.542 | 0.554 | |
| Methyl HT-SELEX, 1 experiments | median | 0.699 | 0.635 | 0.602 | 0.586 | 0.534 | 0.546 |
| best | 0.699 | 0.635 | 0.602 | 0.586 | 0.534 | 0.546 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.714 | 0.652 | 0.616 | 0.597 | 0.542 | 0.554 |
| best | 0.714 | 0.652 | 0.616 | 0.597 | 0.542 | 0.554 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | ZNF148-like {2.3.3.13} (TFClass) |
| TFClass ID | TFClass: 2.3.3.13.2 |
| HGNC | HGNC:13075 |
| MGI | MGI:3029290 |
| EntrezGene (human) | GeneID:23528 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:226442 (SSTAR profile) |
| UniProt ID (human) | ZN281_HUMAN |
| UniProt ID (mouse) | ZN281_MOUSE |
| UniProt AC (human) | Q9Y2X9 (TFClass) |
| UniProt AC (mouse) | Q99LI5 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 6 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| PCM | ZN281.H12INVIVO.1.SM.B.pcm |
| PWM | ZN281.H12INVIVO.1.SM.B.pwm |
| PFM | ZN281.H12INVIVO.1.SM.B.pfm |
| Alignment | ZN281.H12INVIVO.1.SM.B.words.tsv |
| Threshold to P-value map | ZN281.H12INVIVO.1.SM.B.thr |
| Motif in other formats | |
| JASPAR format | ZN281.H12INVIVO.1.SM.B_jaspar_format.txt |
| MEME format | ZN281.H12INVIVO.1.SM.B_meme_format.meme |
| Transfac format | ZN281.H12INVIVO.1.SM.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1228.0 | 892.0 | 3379.0 | 1678.0 |
| 02 | 1134.75 | 570.75 | 3690.75 | 1780.75 |
| 03 | 986.0 | 480.0 | 3711.0 | 2000.0 |
| 04 | 861.0 | 425.0 | 5128.0 | 763.0 |
| 05 | 1108.0 | 575.0 | 4543.0 | 951.0 |
| 06 | 703.0 | 229.0 | 5713.0 | 532.0 |
| 07 | 78.0 | 233.0 | 6620.0 | 246.0 |
| 08 | 678.0 | 91.0 | 6327.0 | 81.0 |
| 09 | 6548.0 | 26.0 | 0.0 | 603.0 |
| 10 | 0.0 | 193.0 | 6814.0 | 170.0 |
| 11 | 2.0 | 6.0 | 7168.0 | 1.0 |
| 12 | 217.0 | 334.0 | 6046.0 | 580.0 |
| 13 | 1721.0 | 657.0 | 3752.0 | 1047.0 |
| 14 | 1843.0 | 1140.0 | 3388.0 | 806.0 |
| 15 | 1432.0 | 535.0 | 4556.0 | 654.0 |
| 16 | 1078.0 | 488.0 | 4763.0 | 848.0 |
| 17 | 978.0 | 589.0 | 4654.0 | 956.0 |
| 18 | 1263.0 | 635.0 | 4309.0 | 970.0 |
| 19 | 1320.0 | 535.0 | 4432.0 | 890.0 |
| 20 | 1194.0 | 585.0 | 4451.0 | 947.0 |
| 21 | 1161.0 | 549.0 | 4625.0 | 842.0 |
| 22 | 1228.0 | 546.0 | 4338.0 | 1065.0 |
| 23 | 1394.0 | 534.0 | 4096.0 | 1153.0 |
| 24 | 1515.0 | 560.0 | 4075.0 | 1027.0 |
| 25 | 1513.75 | 600.75 | 4013.75 | 1048.75 |
| 26 | 1585.75 | 771.75 | 3663.75 | 1155.75 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.171 | 0.124 | 0.471 | 0.234 |
| 02 | 0.158 | 0.08 | 0.514 | 0.248 |
| 03 | 0.137 | 0.067 | 0.517 | 0.279 |
| 04 | 0.12 | 0.059 | 0.715 | 0.106 |
| 05 | 0.154 | 0.08 | 0.633 | 0.133 |
| 06 | 0.098 | 0.032 | 0.796 | 0.074 |
| 07 | 0.011 | 0.032 | 0.922 | 0.034 |
| 08 | 0.094 | 0.013 | 0.882 | 0.011 |
| 09 | 0.912 | 0.004 | 0.0 | 0.084 |
| 10 | 0.0 | 0.027 | 0.949 | 0.024 |
| 11 | 0.0 | 0.001 | 0.999 | 0.0 |
| 12 | 0.03 | 0.047 | 0.842 | 0.081 |
| 13 | 0.24 | 0.092 | 0.523 | 0.146 |
| 14 | 0.257 | 0.159 | 0.472 | 0.112 |
| 15 | 0.2 | 0.075 | 0.635 | 0.091 |
| 16 | 0.15 | 0.068 | 0.664 | 0.118 |
| 17 | 0.136 | 0.082 | 0.648 | 0.133 |
| 18 | 0.176 | 0.088 | 0.6 | 0.135 |
| 19 | 0.184 | 0.075 | 0.618 | 0.124 |
| 20 | 0.166 | 0.082 | 0.62 | 0.132 |
| 21 | 0.162 | 0.076 | 0.644 | 0.117 |
| 22 | 0.171 | 0.076 | 0.604 | 0.148 |
| 23 | 0.194 | 0.074 | 0.571 | 0.161 |
| 24 | 0.211 | 0.078 | 0.568 | 0.143 |
| 25 | 0.211 | 0.084 | 0.559 | 0.146 |
| 26 | 0.221 | 0.108 | 0.51 | 0.161 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.379 | -0.698 | 0.632 | -0.067 |
| 02 | -0.457 | -1.143 | 0.721 | -0.008 |
| 03 | -0.598 | -1.315 | 0.726 | 0.108 |
| 04 | -0.733 | -1.436 | 1.049 | -0.853 |
| 05 | -0.481 | -1.135 | 0.928 | -0.634 |
| 06 | -0.935 | -2.05 | 1.157 | -1.213 |
| 07 | -3.109 | -2.033 | 1.305 | -1.979 |
| 08 | -0.971 | -2.959 | 1.259 | -3.072 |
| 09 | 1.294 | -4.154 | -6.696 | -1.088 |
| 10 | -6.696 | -2.219 | 1.333 | -2.345 |
| 11 | -6.054 | -5.387 | 1.384 | -6.324 |
| 12 | -2.104 | -1.676 | 1.214 | -1.127 |
| 13 | -0.042 | -1.003 | 0.737 | -0.538 |
| 14 | 0.027 | -0.453 | 0.635 | -0.799 |
| 15 | -0.225 | -1.207 | 0.931 | -1.007 |
| 16 | -0.509 | -1.299 | 0.976 | -0.748 |
| 17 | -0.606 | -1.111 | 0.952 | -0.629 |
| 18 | -0.351 | -1.036 | 0.875 | -0.614 |
| 19 | -0.307 | -1.207 | 0.904 | -0.7 |
| 20 | -0.407 | -1.118 | 0.908 | -0.638 |
| 21 | -0.435 | -1.181 | 0.946 | -0.755 |
| 22 | -0.379 | -1.187 | 0.882 | -0.521 |
| 23 | -0.252 | -1.209 | 0.825 | -0.442 |
| 24 | -0.169 | -1.162 | 0.82 | -0.557 |
| 25 | -0.17 | -1.092 | 0.804 | -0.536 |
| 26 | -0.123 | -0.842 | 0.713 | -0.439 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.06146 |
| 0.0005 | 3.29281 |
| 0.0001 | 5.86026 |