| Motif | ZN177.H12INVIVO.0.SM.D |
| Gene (human) | ZNF177 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 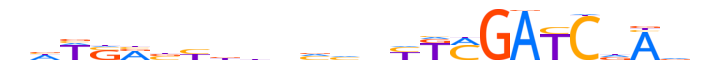 |
| LOGO (reverse complement) | 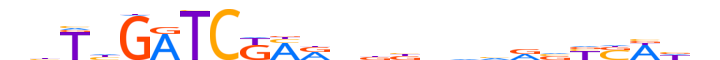 |
| Motif subtype | 0 |
| Quality | D |
| Motif | ZN177.H12INVIVO.0.SM.D |
| Gene (human) | ZNF177 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 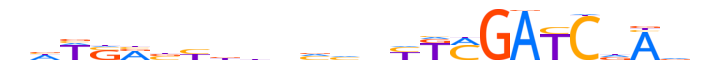 |
| LOGO (reverse complement) | 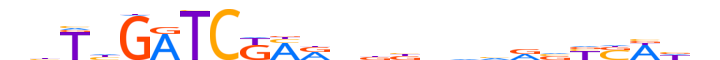 |
| Motif subtype | 0 |
| Quality | D |
| Motif length | 24 |
| Consensus | nhWSRbbdbbvvbYKMGATCdAvn |
| GC content | 48.56% |
| Information content (bits; total / per base) | 13.235 / 0.551 |
| Data sources | HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 9848 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.822 | 0.835 | 0.641 | 0.675 | 0.559 | 0.596 |
| best | 0.889 | 0.893 | 0.683 | 0.714 | 0.58 | 0.62 | |
| Methyl HT-SELEX, 1 experiments | median | 0.754 | 0.776 | 0.599 | 0.636 | 0.537 | 0.572 |
| best | 0.754 | 0.776 | 0.599 | 0.636 | 0.537 | 0.572 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.889 | 0.893 | 0.683 | 0.714 | 0.58 | 0.62 |
| best | 0.889 | 0.893 | 0.683 | 0.714 | 0.58 | 0.62 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | ZNF177-like {2.3.3.40} (TFClass) |
| TFClass ID | TFClass: 2.3.3.40.1 |
| HGNC | HGNC:12966 |
| MGI | |
| EntrezGene (human) | GeneID:100529215 (SSTAR profile) GeneID:7730 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN177_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q13360 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| PCM | ZN177.H12INVIVO.0.SM.D.pcm |
| PWM | ZN177.H12INVIVO.0.SM.D.pwm |
| PFM | ZN177.H12INVIVO.0.SM.D.pfm |
| Alignment | ZN177.H12INVIVO.0.SM.D.words.tsv |
| Threshold to P-value map | ZN177.H12INVIVO.0.SM.D.thr |
| Motif in other formats | |
| JASPAR format | ZN177.H12INVIVO.0.SM.D_jaspar_format.txt |
| MEME format | ZN177.H12INVIVO.0.SM.D_meme_format.meme |
| Transfac format | ZN177.H12INVIVO.0.SM.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 3372.5 | 2791.5 | 1565.5 | 2118.5 |
| 02 | 5509.0 | 1438.0 | 1377.0 | 1524.0 |
| 03 | 1026.0 | 947.0 | 762.0 | 7113.0 |
| 04 | 905.0 | 1704.0 | 6370.0 | 869.0 |
| 05 | 6264.0 | 1094.0 | 1513.0 | 977.0 |
| 06 | 1031.0 | 5003.0 | 2092.0 | 1722.0 |
| 07 | 721.0 | 3661.0 | 739.0 | 4727.0 |
| 08 | 1844.0 | 1187.0 | 2957.0 | 3860.0 |
| 09 | 1467.0 | 1813.0 | 2476.0 | 4092.0 |
| 10 | 1490.0 | 1822.0 | 3447.0 | 3089.0 |
| 11 | 2402.0 | 5027.0 | 1370.0 | 1049.0 |
| 12 | 2550.0 | 4522.0 | 1562.0 | 1214.0 |
| 13 | 1228.0 | 3249.0 | 3242.0 | 2129.0 |
| 14 | 844.0 | 3469.0 | 415.0 | 5120.0 |
| 15 | 670.0 | 446.0 | 1558.0 | 7174.0 |
| 16 | 3077.0 | 6234.0 | 106.0 | 431.0 |
| 17 | 93.0 | 160.0 | 9540.0 | 55.0 |
| 18 | 9442.0 | 154.0 | 226.0 | 26.0 |
| 19 | 88.0 | 2156.0 | 72.0 | 7532.0 |
| 20 | 225.0 | 9219.0 | 90.0 | 314.0 |
| 21 | 2408.0 | 802.0 | 4450.0 | 2188.0 |
| 22 | 8230.0 | 627.0 | 470.0 | 521.0 |
| 23 | 2805.0 | 1935.0 | 3909.0 | 1199.0 |
| 24 | 2426.25 | 2607.25 | 2386.25 | 2428.25 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.342 | 0.283 | 0.159 | 0.215 |
| 02 | 0.559 | 0.146 | 0.14 | 0.155 |
| 03 | 0.104 | 0.096 | 0.077 | 0.722 |
| 04 | 0.092 | 0.173 | 0.647 | 0.088 |
| 05 | 0.636 | 0.111 | 0.154 | 0.099 |
| 06 | 0.105 | 0.508 | 0.212 | 0.175 |
| 07 | 0.073 | 0.372 | 0.075 | 0.48 |
| 08 | 0.187 | 0.121 | 0.3 | 0.392 |
| 09 | 0.149 | 0.184 | 0.251 | 0.416 |
| 10 | 0.151 | 0.185 | 0.35 | 0.314 |
| 11 | 0.244 | 0.51 | 0.139 | 0.107 |
| 12 | 0.259 | 0.459 | 0.159 | 0.123 |
| 13 | 0.125 | 0.33 | 0.329 | 0.216 |
| 14 | 0.086 | 0.352 | 0.042 | 0.52 |
| 15 | 0.068 | 0.045 | 0.158 | 0.728 |
| 16 | 0.312 | 0.633 | 0.011 | 0.044 |
| 17 | 0.009 | 0.016 | 0.969 | 0.006 |
| 18 | 0.959 | 0.016 | 0.023 | 0.003 |
| 19 | 0.009 | 0.219 | 0.007 | 0.765 |
| 20 | 0.023 | 0.936 | 0.009 | 0.032 |
| 21 | 0.245 | 0.081 | 0.452 | 0.222 |
| 22 | 0.836 | 0.064 | 0.048 | 0.053 |
| 23 | 0.285 | 0.196 | 0.397 | 0.122 |
| 24 | 0.246 | 0.265 | 0.242 | 0.247 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.314 | 0.125 | -0.452 | -0.15 |
| 02 | 0.805 | -0.537 | -0.58 | -0.479 |
| 03 | -0.874 | -0.954 | -1.171 | 1.06 |
| 04 | -0.999 | -0.368 | 0.95 | -1.04 |
| 05 | 0.933 | -0.81 | -0.486 | -0.923 |
| 06 | -0.869 | 0.709 | -0.163 | -0.357 |
| 07 | -1.226 | 0.396 | -1.201 | 0.652 |
| 08 | -0.289 | -0.729 | 0.183 | 0.449 |
| 09 | -0.517 | -0.306 | 0.006 | 0.508 |
| 10 | -0.502 | -0.301 | 0.336 | 0.227 |
| 11 | -0.025 | 0.713 | -0.585 | -0.852 |
| 12 | 0.035 | 0.608 | -0.454 | -0.706 |
| 13 | -0.695 | 0.277 | 0.275 | -0.145 |
| 14 | -1.069 | 0.343 | -1.776 | 0.732 |
| 15 | -1.299 | -1.704 | -0.457 | 1.069 |
| 16 | 0.223 | 0.928 | -3.125 | -1.738 |
| 17 | -3.253 | -2.72 | 1.354 | -3.761 |
| 18 | 1.344 | -2.758 | -2.379 | -4.467 |
| 19 | -3.307 | -0.133 | -3.502 | 1.118 |
| 20 | -2.383 | 1.32 | -3.285 | -2.053 |
| 21 | -0.022 | -1.12 | 0.592 | -0.118 |
| 22 | 1.206 | -1.365 | -1.652 | -1.55 |
| 23 | 0.13 | -0.241 | 0.462 | -0.719 |
| 24 | -0.015 | 0.057 | -0.031 | -0.014 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.81876 |
| 0.0005 | 4.82636 |
| 0.0001 | 6.92306 |