| Motif | ZN121.H12INVITRO.0.P.D |
| Gene (human) | ZNF121 (GeneCards) |
| Gene synonyms (human) | ZNF20 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 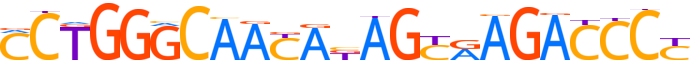 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | D |
| Motif | ZN121.H12INVITRO.0.P.D |
| Gene (human) | ZNF121 (GeneCards) |
| Gene synonyms (human) | ZNF20 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 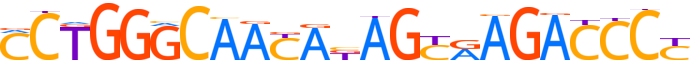 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | D |
| Motif length | 23 |
| Consensus | CCTGGGCAAYAbAGYRAGACCCY |
| GC content | 60.68% |
| Information content (bits; total / per base) | 31.215 / 1.357 |
| Data sources | ChIP-Seq |
| Aligned words | 869 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 2 (11) | 0.884 | 0.955 | 0.858 | 0.931 | 0.924 | 0.984 | 16.084 | 18.819 | 200.292 | 304.854 |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 1.889 | 0.536 | 0.112 | 0.052 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | Unclassified {2.3.3.0} (TFClass) |
| TFClass ID | TFClass: 2.3.3.0.112 |
| HGNC | HGNC:12904 |
| MGI | |
| EntrezGene (human) | GeneID:7675 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN121_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | P58317 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 0 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | ZN121.H12INVITRO.0.P.D.pcm |
| PWM | ZN121.H12INVITRO.0.P.D.pwm |
| PFM | ZN121.H12INVITRO.0.P.D.pfm |
| Alignment | ZN121.H12INVITRO.0.P.D.words.tsv |
| Threshold to P-value map | ZN121.H12INVITRO.0.P.D.thr |
| Motif in other formats | |
| JASPAR format | ZN121.H12INVITRO.0.P.D_jaspar_format.txt |
| MEME format | ZN121.H12INVITRO.0.P.D_meme_format.meme |
| Transfac format | ZN121.H12INVITRO.0.P.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 87.0 | 717.0 | 45.0 | 20.0 |
| 02 | 10.0 | 824.0 | 6.0 | 29.0 |
| 03 | 42.0 | 12.0 | 60.0 | 755.0 |
| 04 | 2.0 | 2.0 | 860.0 | 5.0 |
| 05 | 14.0 | 11.0 | 840.0 | 4.0 |
| 06 | 70.0 | 44.0 | 753.0 | 2.0 |
| 07 | 9.0 | 851.0 | 7.0 | 2.0 |
| 08 | 800.0 | 42.0 | 12.0 | 15.0 |
| 09 | 768.0 | 19.0 | 71.0 | 11.0 |
| 10 | 28.0 | 676.0 | 56.0 | 109.0 |
| 11 | 743.0 | 19.0 | 85.0 | 22.0 |
| 12 | 14.0 | 199.0 | 199.0 | 457.0 |
| 13 | 801.0 | 13.0 | 15.0 | 40.0 |
| 14 | 6.0 | 8.0 | 845.0 | 10.0 |
| 15 | 16.0 | 535.0 | 18.0 | 300.0 |
| 16 | 459.0 | 35.0 | 364.0 | 11.0 |
| 17 | 814.0 | 19.0 | 21.0 | 15.0 |
| 18 | 9.0 | 8.0 | 848.0 | 4.0 |
| 19 | 818.0 | 10.0 | 31.0 | 10.0 |
| 20 | 5.0 | 756.0 | 10.0 | 98.0 |
| 21 | 11.0 | 782.0 | 11.0 | 65.0 |
| 22 | 8.0 | 837.0 | 6.0 | 18.0 |
| 23 | 29.0 | 530.0 | 16.0 | 294.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.1 | 0.825 | 0.052 | 0.023 |
| 02 | 0.012 | 0.948 | 0.007 | 0.033 |
| 03 | 0.048 | 0.014 | 0.069 | 0.869 |
| 04 | 0.002 | 0.002 | 0.99 | 0.006 |
| 05 | 0.016 | 0.013 | 0.967 | 0.005 |
| 06 | 0.081 | 0.051 | 0.867 | 0.002 |
| 07 | 0.01 | 0.979 | 0.008 | 0.002 |
| 08 | 0.921 | 0.048 | 0.014 | 0.017 |
| 09 | 0.884 | 0.022 | 0.082 | 0.013 |
| 10 | 0.032 | 0.778 | 0.064 | 0.125 |
| 11 | 0.855 | 0.022 | 0.098 | 0.025 |
| 12 | 0.016 | 0.229 | 0.229 | 0.526 |
| 13 | 0.922 | 0.015 | 0.017 | 0.046 |
| 14 | 0.007 | 0.009 | 0.972 | 0.012 |
| 15 | 0.018 | 0.616 | 0.021 | 0.345 |
| 16 | 0.528 | 0.04 | 0.419 | 0.013 |
| 17 | 0.937 | 0.022 | 0.024 | 0.017 |
| 18 | 0.01 | 0.009 | 0.976 | 0.005 |
| 19 | 0.941 | 0.012 | 0.036 | 0.012 |
| 20 | 0.006 | 0.87 | 0.012 | 0.113 |
| 21 | 0.013 | 0.9 | 0.013 | 0.075 |
| 22 | 0.009 | 0.963 | 0.007 | 0.021 |
| 23 | 0.033 | 0.61 | 0.018 | 0.338 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.904 | 1.189 | -1.545 | -2.312 |
| 02 | -2.93 | 1.327 | -3.349 | -1.965 |
| 03 | -1.612 | -2.772 | -1.267 | 1.24 |
| 04 | -4.083 | -4.083 | 1.37 | -3.488 |
| 05 | -2.636 | -2.848 | 1.347 | -3.65 |
| 06 | -1.116 | -1.567 | 1.238 | -4.083 |
| 07 | -3.019 | 1.36 | -3.226 | -4.083 |
| 08 | 1.298 | -1.612 | -2.772 | -2.574 |
| 09 | 1.257 | -2.359 | -1.103 | -2.848 |
| 10 | -1.998 | 1.13 | -1.334 | -0.682 |
| 11 | 1.224 | -2.359 | -0.926 | -2.224 |
| 12 | -2.636 | -0.087 | -0.087 | 0.74 |
| 13 | 1.299 | -2.702 | -2.574 | -1.659 |
| 14 | -3.349 | -3.118 | 1.353 | -2.93 |
| 15 | -2.516 | 0.897 | -2.409 | 0.321 |
| 16 | 0.744 | -1.786 | 0.513 | -2.848 |
| 17 | 1.315 | -2.359 | -2.267 | -2.574 |
| 18 | -3.019 | -3.118 | 1.356 | -3.65 |
| 19 | 1.32 | -2.93 | -1.902 | -2.93 |
| 20 | -3.488 | 1.241 | -2.93 | -0.787 |
| 21 | -2.848 | 1.275 | -2.848 | -1.189 |
| 22 | -3.118 | 1.343 | -3.349 | -2.409 |
| 23 | -1.965 | 0.887 | -2.516 | 0.301 |
| P-value | Threshold |
|---|---|
| 0.001 | -6.11229 |
| 0.0005 | -4.35754 |
| 0.0001 | -0.58519 |