| Motif | ZF64B.H12INVIVO.0.P.B |
| Gene (human) | ZFP64 (GeneCards) |
| Gene synonyms (human) | ZNF338 |
| Gene (mouse) | Zfp64 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 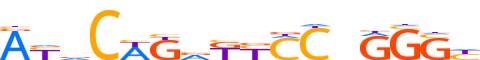 |
| Motif subtype | 0 |
| Quality | B |
| Motif | ZF64B.H12INVIVO.0.P.B |
| Gene (human) | ZFP64 (GeneCards) |
| Gene synonyms (human) | ZNF338 |
| Gene (mouse) | Zfp64 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 16 |
| Consensus | dSCYnGKRMbMWGbRT |
| GC content | 59.24% |
| Information content (bits; total / per base) | 12.336 / 0.771 |
| Data sources | ChIP-Seq |
| Aligned words | 830 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | 2 (14) | 0.878 | 0.89 | 0.839 | 0.857 | 0.884 | 0.892 | 4.763 | 4.935 | 227.602 | 278.721 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | Unclassified {2.3.3.0} (TFClass) |
| TFClass ID | TFClass: 2.3.3.0.76 |
| HGNC | HGNC:15940 |
| MGI | MGI:107342 |
| EntrezGene (human) | GeneID:55734 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZF64B_HUMAN |
| UniProt ID (mouse) | ZFP64_MOUSE |
| UniProt AC (human) | Q9NTW7 (TFClass) |
| UniProt AC (mouse) | Q99KE8 (TFClass) |
| GRECO-DB-TF | no |
| ChIP-Seq | 0 human, 2 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | ZF64B.H12INVIVO.0.P.B.pcm |
| PWM | ZF64B.H12INVIVO.0.P.B.pwm |
| PFM | ZF64B.H12INVIVO.0.P.B.pfm |
| Alignment | ZF64B.H12INVIVO.0.P.B.words.tsv |
| Threshold to P-value map | ZF64B.H12INVIVO.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | ZF64B.H12INVIVO.0.P.B_jaspar_format.txt |
| MEME format | ZF64B.H12INVIVO.0.P.B_meme_format.meme |
| Transfac format | ZF64B.H12INVIVO.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 255.0 | 54.0 | 437.0 | 84.0 |
| 02 | 33.0 | 646.0 | 112.0 | 39.0 |
| 03 | 27.0 | 710.0 | 59.0 | 34.0 |
| 04 | 51.0 | 667.0 | 40.0 | 72.0 |
| 05 | 234.0 | 160.0 | 261.0 | 175.0 |
| 06 | 54.0 | 13.0 | 717.0 | 46.0 |
| 07 | 58.0 | 46.0 | 655.0 | 71.0 |
| 08 | 515.0 | 105.0 | 197.0 | 13.0 |
| 09 | 496.0 | 255.0 | 68.0 | 11.0 |
| 10 | 86.0 | 190.0 | 112.0 | 442.0 |
| 11 | 106.0 | 621.0 | 91.0 | 12.0 |
| 12 | 188.0 | 12.0 | 91.0 | 539.0 |
| 13 | 10.0 | 13.0 | 799.0 | 8.0 |
| 14 | 113.0 | 281.0 | 123.0 | 313.0 |
| 15 | 529.0 | 66.0 | 173.0 | 62.0 |
| 16 | 35.0 | 49.0 | 44.0 | 702.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.307 | 0.065 | 0.527 | 0.101 |
| 02 | 0.04 | 0.778 | 0.135 | 0.047 |
| 03 | 0.033 | 0.855 | 0.071 | 0.041 |
| 04 | 0.061 | 0.804 | 0.048 | 0.087 |
| 05 | 0.282 | 0.193 | 0.314 | 0.211 |
| 06 | 0.065 | 0.016 | 0.864 | 0.055 |
| 07 | 0.07 | 0.055 | 0.789 | 0.086 |
| 08 | 0.62 | 0.127 | 0.237 | 0.016 |
| 09 | 0.598 | 0.307 | 0.082 | 0.013 |
| 10 | 0.104 | 0.229 | 0.135 | 0.533 |
| 11 | 0.128 | 0.748 | 0.11 | 0.014 |
| 12 | 0.227 | 0.014 | 0.11 | 0.649 |
| 13 | 0.012 | 0.016 | 0.963 | 0.01 |
| 14 | 0.136 | 0.339 | 0.148 | 0.377 |
| 15 | 0.637 | 0.08 | 0.208 | 0.075 |
| 16 | 0.042 | 0.059 | 0.053 | 0.846 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.205 | -1.324 | 0.741 | -0.893 |
| 02 | -1.797 | 1.13 | -0.61 | -1.637 |
| 03 | -1.987 | 1.224 | -1.238 | -1.769 |
| 04 | -1.379 | 1.162 | -1.613 | -1.043 |
| 05 | 0.119 | -0.258 | 0.228 | -0.169 |
| 06 | -1.324 | -2.657 | 1.234 | -1.479 |
| 07 | -1.254 | -1.479 | 1.144 | -1.057 |
| 08 | 0.904 | -0.673 | -0.051 | -2.657 |
| 09 | 0.867 | 0.205 | -1.099 | -2.803 |
| 10 | -0.869 | -0.087 | -0.61 | 0.752 |
| 11 | -0.664 | 1.091 | -0.814 | -2.727 |
| 12 | -0.098 | -2.727 | -0.814 | 0.95 |
| 13 | -2.885 | -2.657 | 1.342 | -3.073 |
| 14 | -0.601 | 0.301 | -0.517 | 0.408 |
| 15 | 0.931 | -1.128 | -0.18 | -1.189 |
| 16 | -1.741 | -1.418 | -1.522 | 1.213 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.17476 |
| 0.0005 | 5.08831 |
| 0.0001 | 7.01186 |