| Motif | VENTX.H12INVITRO.0.SM.B |
| Gene (human) | VENTX (GeneCards) |
| Gene synonyms (human) | HPX42B, VENTX2 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 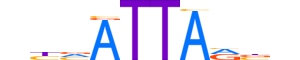 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | VENTX.H12INVITRO.0.SM.B |
| Gene (human) | VENTX (GeneCards) |
| Gene synonyms (human) | HPX42B, VENTX2 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 10 |
| Consensus | nhvATTARbn |
| GC content | 35.91% |
| Information content (bits; total / per base) | 8.493 / 0.849 |
| Data sources | HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 9580 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.953 | 0.933 | 0.905 | 0.877 | 0.858 | 0.827 |
| best | 0.987 | 0.981 | 0.97 | 0.957 | 0.941 | 0.92 | |
| Methyl HT-SELEX, 1 experiments | median | 0.987 | 0.981 | 0.97 | 0.957 | 0.941 | 0.92 |
| best | 0.987 | 0.981 | 0.97 | 0.957 | 0.941 | 0.92 | |
| Non-Methyl HT-SELEX, 3 experiments | median | 0.928 | 0.9 | 0.852 | 0.817 | 0.787 | 0.752 |
| best | 0.978 | 0.966 | 0.958 | 0.938 | 0.929 | 0.902 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.935 | 0.138 | 0.915 | 0.52 |
| batch 2 | 0.783 | 0.208 | 0.752 | 0.49 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | NK-related {3.1.2} (TFClass) |
| TF subfamily | VENTX {3.1.2.23} (TFClass) |
| TFClass ID | TFClass: 3.1.2.23.1 |
| HGNC | HGNC:13639 |
| MGI | |
| EntrezGene (human) | GeneID:27287 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | VENTX_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | O95231 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 3 |
| Methyl-HT-SELEX | 1 |
| PCM | VENTX.H12INVITRO.0.SM.B.pcm |
| PWM | VENTX.H12INVITRO.0.SM.B.pwm |
| PFM | VENTX.H12INVITRO.0.SM.B.pfm |
| Alignment | VENTX.H12INVITRO.0.SM.B.words.tsv |
| Threshold to P-value map | VENTX.H12INVITRO.0.SM.B.thr |
| Motif in other formats | |
| JASPAR format | VENTX.H12INVITRO.0.SM.B_jaspar_format.txt |
| MEME format | VENTX.H12INVITRO.0.SM.B_meme_format.meme |
| Transfac format | VENTX.H12INVITRO.0.SM.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 2378.5 | 2798.5 | 2390.5 | 2012.5 |
| 02 | 883.5 | 4115.5 | 766.5 | 3814.5 |
| 03 | 3139.0 | 3754.0 | 2417.0 | 270.0 |
| 04 | 8772.0 | 350.0 | 394.0 | 64.0 |
| 05 | 0.0 | 1.0 | 0.0 | 9579.0 |
| 06 | 0.0 | 2.0 | 1.0 | 9577.0 |
| 07 | 9145.0 | 386.0 | 27.0 | 22.0 |
| 08 | 3644.0 | 796.0 | 4555.0 | 585.0 |
| 09 | 784.25 | 4483.25 | 2224.25 | 2088.25 |
| 10 | 2709.75 | 1803.75 | 3138.75 | 1927.75 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.248 | 0.292 | 0.25 | 0.21 |
| 02 | 0.092 | 0.43 | 0.08 | 0.398 |
| 03 | 0.328 | 0.392 | 0.252 | 0.028 |
| 04 | 0.916 | 0.037 | 0.041 | 0.007 |
| 05 | 0.0 | 0.0 | 0.0 | 1.0 |
| 06 | 0.0 | 0.0 | 0.0 | 1.0 |
| 07 | 0.955 | 0.04 | 0.003 | 0.002 |
| 08 | 0.38 | 0.083 | 0.475 | 0.061 |
| 09 | 0.082 | 0.468 | 0.232 | 0.218 |
| 10 | 0.283 | 0.188 | 0.328 | 0.201 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.007 | 0.156 | -0.002 | -0.174 |
| 02 | -0.996 | 0.541 | -1.137 | 0.465 |
| 03 | 0.27 | 0.449 | 0.009 | -2.175 |
| 04 | 1.297 | -1.918 | -1.8 | -3.588 |
| 05 | -6.953 | -6.591 | -6.953 | 1.385 |
| 06 | -6.953 | -6.325 | -6.591 | 1.385 |
| 07 | 1.339 | -1.82 | -4.405 | -4.592 |
| 08 | 0.419 | -1.1 | 0.642 | -1.407 |
| 09 | -1.114 | 0.627 | -0.074 | -0.137 |
| 10 | 0.123 | -0.283 | 0.27 | -0.217 |
| P-value | Threshold |
|---|---|
| 0.001 | 5.383985 |
| 0.0005 | 6.22016 |
| 0.0001 | 7.25935 |