| Motif | UBIP1.H12INVITRO.0.SM.B |
| Gene (human) | UBP1 (GeneCards) |
| Gene synonyms (human) | LBP1 |
| Gene (mouse) | Ubp1 |
| Gene synonyms (mouse) | Cp2b, Nf2d9 |
| LOGO | 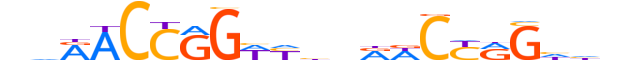 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | UBIP1.H12INVITRO.0.SM.B |
| Gene (human) | UBP1 (GeneCards) |
| Gene synonyms (human) | LBP1 |
| Gene (mouse) | Ubp1 |
| Gene synonyms (mouse) | Cp2b, Nf2d9 |
| LOGO | 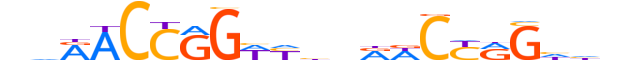 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 21 |
| Consensus | ndRACCGGWYhvRWCYRGhhn |
| GC content | 51.92% |
| Information content (bits; total / per base) | 15.103 / 0.719 |
| Data sources | HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 9804 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.998 | 0.996 | 0.993 | 0.99 | 0.979 | 0.972 |
| best | 0.999 | 0.998 | 0.997 | 0.995 | 0.991 | 0.988 | |
| Methyl HT-SELEX, 2 experiments | median | 0.998 | 0.997 | 0.994 | 0.991 | 0.979 | 0.973 |
| best | 0.999 | 0.998 | 0.997 | 0.995 | 0.991 | 0.988 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.997 | 0.995 | 0.993 | 0.99 | 0.979 | 0.972 |
| best | 0.998 | 0.997 | 0.994 | 0.992 | 0.982 | 0.975 | |
| TF superclass | Immunoglobulin fold {6} (TFClass) |
| TF class | Grainyhead domain factors {6.7} (TFClass) |
| TF family | CP2-related {6.7.2} (TFClass) |
| TF subfamily | {6.7.2.0} (TFClass) |
| TFClass ID | TFClass: 6.7.2.0.3 |
| HGNC | HGNC:12507 |
| MGI | MGI:104889 |
| EntrezGene (human) | GeneID:7342 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:22221 (SSTAR profile) |
| UniProt ID (human) | UBIP1_HUMAN |
| UniProt ID (mouse) | UBIP1_MOUSE |
| UniProt AC (human) | Q9NZI7 (TFClass) |
| UniProt AC (mouse) | Q811S7 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 2 |
| PCM | UBIP1.H12INVITRO.0.SM.B.pcm |
| PWM | UBIP1.H12INVITRO.0.SM.B.pwm |
| PFM | UBIP1.H12INVITRO.0.SM.B.pfm |
| Alignment | UBIP1.H12INVITRO.0.SM.B.words.tsv |
| Threshold to P-value map | UBIP1.H12INVITRO.0.SM.B.thr |
| Motif in other formats | |
| JASPAR format | UBIP1.H12INVITRO.0.SM.B_jaspar_format.txt |
| MEME format | UBIP1.H12INVITRO.0.SM.B_meme_format.meme |
| Transfac format | UBIP1.H12INVITRO.0.SM.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1710.0 | 2231.0 | 3520.0 | 2343.0 |
| 02 | 3801.5 | 1480.5 | 2830.5 | 1691.5 |
| 03 | 6876.0 | 329.0 | 1315.0 | 1284.0 |
| 04 | 7846.0 | 2.0 | 249.0 | 1707.0 |
| 05 | 0.0 | 9774.0 | 28.0 | 2.0 |
| 06 | 43.0 | 7973.0 | 4.0 | 1784.0 |
| 07 | 2479.0 | 23.0 | 7264.0 | 38.0 |
| 08 | 22.0 | 349.0 | 9432.0 | 1.0 |
| 09 | 2864.0 | 1355.0 | 23.0 | 5562.0 |
| 10 | 1634.0 | 1669.0 | 442.0 | 6059.0 |
| 11 | 1738.0 | 3327.0 | 1316.0 | 3423.0 |
| 12 | 3747.0 | 1697.0 | 2685.0 | 1675.0 |
| 13 | 6146.0 | 636.0 | 1688.0 | 1334.0 |
| 14 | 5838.0 | 257.0 | 1746.0 | 1963.0 |
| 15 | 34.0 | 9145.0 | 459.0 | 166.0 |
| 16 | 609.0 | 6225.0 | 206.0 | 2764.0 |
| 17 | 3270.0 | 261.0 | 5625.0 | 648.0 |
| 18 | 301.0 | 944.0 | 8527.0 | 32.0 |
| 19 | 3151.0 | 2968.0 | 287.0 | 3398.0 |
| 20 | 2129.25 | 2506.25 | 961.25 | 4207.25 |
| 21 | 2280.5 | 2970.5 | 2165.5 | 2387.5 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.174 | 0.228 | 0.359 | 0.239 |
| 02 | 0.388 | 0.151 | 0.289 | 0.173 |
| 03 | 0.701 | 0.034 | 0.134 | 0.131 |
| 04 | 0.8 | 0.0 | 0.025 | 0.174 |
| 05 | 0.0 | 0.997 | 0.003 | 0.0 |
| 06 | 0.004 | 0.813 | 0.0 | 0.182 |
| 07 | 0.253 | 0.002 | 0.741 | 0.004 |
| 08 | 0.002 | 0.036 | 0.962 | 0.0 |
| 09 | 0.292 | 0.138 | 0.002 | 0.567 |
| 10 | 0.167 | 0.17 | 0.045 | 0.618 |
| 11 | 0.177 | 0.339 | 0.134 | 0.349 |
| 12 | 0.382 | 0.173 | 0.274 | 0.171 |
| 13 | 0.627 | 0.065 | 0.172 | 0.136 |
| 14 | 0.595 | 0.026 | 0.178 | 0.2 |
| 15 | 0.003 | 0.933 | 0.047 | 0.017 |
| 16 | 0.062 | 0.635 | 0.021 | 0.282 |
| 17 | 0.334 | 0.027 | 0.574 | 0.066 |
| 18 | 0.031 | 0.096 | 0.87 | 0.003 |
| 19 | 0.321 | 0.303 | 0.029 | 0.347 |
| 20 | 0.217 | 0.256 | 0.098 | 0.429 |
| 21 | 0.233 | 0.303 | 0.221 | 0.244 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.36 | -0.094 | 0.362 | -0.045 |
| 02 | 0.439 | -0.504 | 0.144 | -0.37 |
| 03 | 1.031 | -2.002 | -0.622 | -0.646 |
| 04 | 1.163 | -6.347 | -2.279 | -0.361 |
| 05 | -6.973 | 1.383 | -4.394 | -6.347 |
| 06 | -3.992 | 1.179 | -5.965 | -0.317 |
| 07 | 0.011 | -4.574 | 1.086 | -4.109 |
| 08 | -4.615 | -1.944 | 1.347 | -6.612 |
| 09 | 0.156 | -0.592 | -4.574 | 0.819 |
| 10 | -0.405 | -0.384 | -1.709 | 0.904 |
| 11 | -0.343 | 0.305 | -0.621 | 0.334 |
| 12 | 0.424 | -0.367 | 0.091 | -0.38 |
| 13 | 0.919 | -1.346 | -0.373 | -0.608 |
| 14 | 0.867 | -2.247 | -0.339 | -0.222 |
| 15 | -4.213 | 1.316 | -1.671 | -2.679 |
| 16 | -1.39 | 0.932 | -2.466 | 0.12 |
| 17 | 0.288 | -2.232 | 0.83 | -1.328 |
| 18 | -2.09 | -0.953 | 1.246 | -4.27 |
| 19 | 0.251 | 0.191 | -2.138 | 0.326 |
| 20 | -0.141 | 0.022 | -0.935 | 0.54 |
| 21 | -0.072 | 0.192 | -0.124 | -0.026 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.59091 |
| 0.0005 | 3.89816 |
| 0.0001 | 6.53696 |