| Motif | TGIF2.H12INVITRO.0.S.B |
| Gene (human) | TGIF2 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Tgif2 |
| Gene synonyms (mouse) | |
| LOGO | 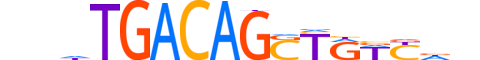 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | TGIF2.H12INVITRO.0.S.B |
| Gene (human) | TGIF2 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Tgif2 |
| Gene synonyms (mouse) | |
| LOGO | 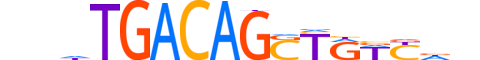 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 16 |
| Consensus | nnhTGACAGSTRWYdn |
| GC content | 48.58% |
| Information content (bits; total / per base) | 16.022 / 1.001 |
| Data sources | HT-SELEX |
| Aligned words | 2499 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (7) | 0.69 | 0.752 | 0.539 | 0.607 | 0.634 | 0.679 | 1.722 | 1.852 | 51.42 | 83.921 |
| Mouse | 1 (6) | 0.886 | 0.887 | 0.807 | 0.809 | 0.876 | 0.892 | 3.343 | 3.416 | 76.638 | 169.699 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 6 experiments | median | 0.974 | 0.958 | 0.951 | 0.933 | 0.889 | 0.853 |
| best | 0.995 | 0.994 | 0.98 | 0.968 | 0.964 | 0.947 | |
| Methyl HT-SELEX, 2 experiments | median | 0.974 | 0.958 | 0.96 | 0.938 | 0.932 | 0.906 |
| best | 0.986 | 0.977 | 0.977 | 0.964 | 0.959 | 0.941 | |
| Non-Methyl HT-SELEX, 4 experiments | median | 0.968 | 0.948 | 0.941 | 0.919 | 0.846 | 0.832 |
| best | 0.995 | 0.994 | 0.98 | 0.968 | 0.964 | 0.947 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.815 | 0.604 | 0.771 | 0.559 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | TALE-type HD {3.1.4} (TFClass) |
| TF subfamily | TGIF {3.1.4.6} (TFClass) |
| TFClass ID | TFClass: 3.1.4.6.2 |
| HGNC | HGNC:15764 |
| MGI | MGI:1915299 |
| EntrezGene (human) | GeneID:60436 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:228839 (SSTAR profile) |
| UniProt ID (human) | TGIF2_HUMAN |
| UniProt ID (mouse) | TGIF2_MOUSE |
| UniProt AC (human) | Q9GZN2 (TFClass) |
| UniProt AC (mouse) | Q8C0Y1 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 1 mouse |
| HT-SELEX | 4 |
| Methyl-HT-SELEX | 2 |
| PCM | TGIF2.H12INVITRO.0.S.B.pcm |
| PWM | TGIF2.H12INVITRO.0.S.B.pwm |
| PFM | TGIF2.H12INVITRO.0.S.B.pfm |
| Alignment | TGIF2.H12INVITRO.0.S.B.words.tsv |
| Threshold to P-value map | TGIF2.H12INVITRO.0.S.B.thr |
| Motif in other formats | |
| JASPAR format | TGIF2.H12INVITRO.0.S.B_jaspar_format.txt |
| MEME format | TGIF2.H12INVITRO.0.S.B_meme_format.meme |
| Transfac format | TGIF2.H12INVITRO.0.S.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 816.0 | 517.0 | 645.0 | 521.0 |
| 02 | 820.25 | 414.25 | 569.25 | 695.25 |
| 03 | 897.0 | 343.0 | 269.0 | 990.0 |
| 04 | 19.0 | 8.0 | 6.0 | 2466.0 |
| 05 | 0.0 | 0.0 | 2499.0 | 0.0 |
| 06 | 2472.0 | 0.0 | 6.0 | 21.0 |
| 07 | 2.0 | 2497.0 | 0.0 | 0.0 |
| 08 | 2493.0 | 1.0 | 5.0 | 0.0 |
| 09 | 3.0 | 36.0 | 2394.0 | 66.0 |
| 10 | 114.0 | 1870.0 | 427.0 | 88.0 |
| 11 | 133.0 | 191.0 | 114.0 | 2061.0 |
| 12 | 270.0 | 151.0 | 1866.0 | 212.0 |
| 13 | 308.0 | 231.0 | 241.0 | 1719.0 |
| 14 | 170.0 | 1790.0 | 222.0 | 317.0 |
| 15 | 1296.75 | 358.75 | 402.75 | 440.75 |
| 16 | 534.75 | 759.75 | 591.75 | 612.75 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.327 | 0.207 | 0.258 | 0.208 |
| 02 | 0.328 | 0.166 | 0.228 | 0.278 |
| 03 | 0.359 | 0.137 | 0.108 | 0.396 |
| 04 | 0.008 | 0.003 | 0.002 | 0.987 |
| 05 | 0.0 | 0.0 | 1.0 | 0.0 |
| 06 | 0.989 | 0.0 | 0.002 | 0.008 |
| 07 | 0.001 | 0.999 | 0.0 | 0.0 |
| 08 | 0.998 | 0.0 | 0.002 | 0.0 |
| 09 | 0.001 | 0.014 | 0.958 | 0.026 |
| 10 | 0.046 | 0.748 | 0.171 | 0.035 |
| 11 | 0.053 | 0.076 | 0.046 | 0.825 |
| 12 | 0.108 | 0.06 | 0.747 | 0.085 |
| 13 | 0.123 | 0.092 | 0.096 | 0.688 |
| 14 | 0.068 | 0.716 | 0.089 | 0.127 |
| 15 | 0.519 | 0.144 | 0.161 | 0.176 |
| 16 | 0.214 | 0.304 | 0.237 | 0.245 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.266 | -0.189 | 0.032 | -0.181 |
| 02 | 0.272 | -0.409 | -0.093 | 0.107 |
| 03 | 0.361 | -0.597 | -0.839 | 0.459 |
| 04 | -3.398 | -4.142 | -4.367 | 1.371 |
| 05 | -5.77 | -5.77 | 1.384 | -5.77 |
| 06 | 1.373 | -5.77 | -4.367 | -3.307 |
| 07 | -5.065 | 1.383 | -5.77 | -5.77 |
| 08 | 1.382 | -5.357 | -4.501 | -5.77 |
| 09 | -4.84 | -2.804 | 1.341 | -2.222 |
| 10 | -1.687 | 1.094 | -0.379 | -1.941 |
| 11 | -1.536 | -1.178 | -1.687 | 1.191 |
| 12 | -0.835 | -1.41 | 1.092 | -1.075 |
| 13 | -0.704 | -0.99 | -0.948 | 1.01 |
| 14 | -1.293 | 1.051 | -1.029 | -0.675 |
| 15 | 0.729 | -0.552 | -0.437 | -0.348 |
| 16 | -0.155 | 0.195 | -0.054 | -0.019 |
| P-value | Threshold |
|---|---|
| 0.001 | 1.53436 |
| 0.0005 | 3.00851 |
| 0.0001 | 6.09661 |