| Motif | TFCP2.H12INVITRO.1.SM.B |
| Gene (human) | TFCP2 (GeneCards) |
| Gene synonyms (human) | LSF, SEF |
| Gene (mouse) | Tfcp2 |
| Gene synonyms (mouse) | Tcfcp2 |
| LOGO | 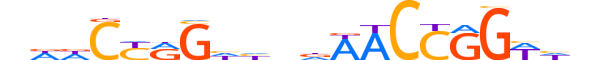 |
| LOGO (reverse complement) | 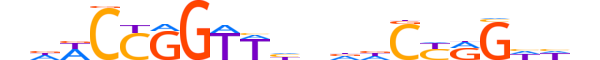 |
| Motif subtype | 1 |
| Quality | B |
| Motif | TFCP2.H12INVITRO.1.SM.B |
| Gene (human) | TFCP2 (GeneCards) |
| Gene synonyms (human) | LSF, SEF |
| Gene (mouse) | Tfcp2 |
| Gene synonyms (mouse) | Tcfcp2 |
| LOGO | 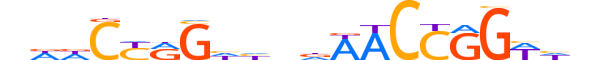 |
| LOGO (reverse complement) | 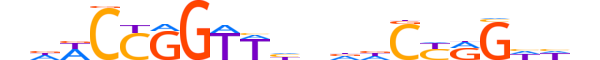 |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 20 |
| Consensus | nddCYRGhhndWACCGGWhn |
| GC content | 50.36% |
| Information content (bits; total / per base) | 14.993 / 0.75 |
| Data sources | HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 9816 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 5 experiments | median | 0.992 | 0.987 | 0.976 | 0.967 | 0.928 | 0.914 |
| best | 1.0 | 0.999 | 0.999 | 0.999 | 0.997 | 0.996 | |
| Methyl HT-SELEX, 2 experiments | median | 0.996 | 0.993 | 0.988 | 0.982 | 0.962 | 0.954 |
| best | 0.999 | 0.999 | 0.999 | 0.998 | 0.996 | 0.995 | |
| Non-Methyl HT-SELEX, 3 experiments | median | 0.96 | 0.953 | 0.78 | 0.792 | 0.635 | 0.672 |
| best | 1.0 | 0.999 | 0.999 | 0.999 | 0.997 | 0.996 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.902 | 0.752 | 0.885 | 0.588 |
| batch 2 | 0.707 | 0.196 | 0.767 | 0.525 |
| TF superclass | Immunoglobulin fold {6} (TFClass) |
| TF class | Grainyhead domain factors {6.7} (TFClass) |
| TF family | CP2-related {6.7.2} (TFClass) |
| TF subfamily | {6.7.2.0} (TFClass) |
| TFClass ID | TFClass: 6.7.2.0.1 |
| HGNC | HGNC:11748 |
| MGI | MGI:98509 |
| EntrezGene (human) | GeneID:7024 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:21422 (SSTAR profile) |
| UniProt ID (human) | TFCP2_HUMAN |
| UniProt ID (mouse) | TFCP2_MOUSE |
| UniProt AC (human) | Q12800 (TFClass) |
| UniProt AC (mouse) | Q9ERA0 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 3 |
| Methyl-HT-SELEX | 2 |
| PCM | TFCP2.H12INVITRO.1.SM.B.pcm |
| PWM | TFCP2.H12INVITRO.1.SM.B.pwm |
| PFM | TFCP2.H12INVITRO.1.SM.B.pfm |
| Alignment | TFCP2.H12INVITRO.1.SM.B.words.tsv |
| Threshold to P-value map | TFCP2.H12INVITRO.1.SM.B.thr |
| Motif in other formats | |
| JASPAR format | TFCP2.H12INVITRO.1.SM.B_jaspar_format.txt |
| MEME format | TFCP2.H12INVITRO.1.SM.B_meme_format.meme |
| Transfac format | TFCP2.H12INVITRO.1.SM.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 3294.25 | 1905.25 | 2589.25 | 2027.25 |
| 02 | 5663.25 | 758.25 | 1646.25 | 1748.25 |
| 03 | 5061.0 | 312.0 | 1822.0 | 2621.0 |
| 04 | 63.0 | 8837.0 | 676.0 | 240.0 |
| 05 | 618.0 | 6134.0 | 303.0 | 2761.0 |
| 06 | 3080.0 | 309.0 | 5812.0 | 615.0 |
| 07 | 250.0 | 810.0 | 8689.0 | 67.0 |
| 08 | 2737.0 | 2593.0 | 279.0 | 4207.0 |
| 09 | 1710.0 | 2421.0 | 902.0 | 4783.0 |
| 10 | 2226.0 | 2271.0 | 2060.0 | 3259.0 |
| 11 | 4354.0 | 779.0 | 3237.0 | 1446.0 |
| 12 | 7480.0 | 145.0 | 821.0 | 1370.0 |
| 13 | 7552.0 | 5.0 | 108.0 | 2151.0 |
| 14 | 1.0 | 9809.0 | 6.0 | 0.0 |
| 15 | 35.0 | 7795.0 | 13.0 | 1973.0 |
| 16 | 2189.0 | 0.0 | 7572.0 | 55.0 |
| 17 | 8.0 | 288.0 | 9519.0 | 1.0 |
| 18 | 3075.0 | 962.0 | 50.0 | 5729.0 |
| 19 | 2198.5 | 1700.5 | 652.5 | 5264.5 |
| 20 | 2348.0 | 2581.0 | 1695.0 | 3192.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.336 | 0.194 | 0.264 | 0.207 |
| 02 | 0.577 | 0.077 | 0.168 | 0.178 |
| 03 | 0.516 | 0.032 | 0.186 | 0.267 |
| 04 | 0.006 | 0.9 | 0.069 | 0.024 |
| 05 | 0.063 | 0.625 | 0.031 | 0.281 |
| 06 | 0.314 | 0.031 | 0.592 | 0.063 |
| 07 | 0.025 | 0.083 | 0.885 | 0.007 |
| 08 | 0.279 | 0.264 | 0.028 | 0.429 |
| 09 | 0.174 | 0.247 | 0.092 | 0.487 |
| 10 | 0.227 | 0.231 | 0.21 | 0.332 |
| 11 | 0.444 | 0.079 | 0.33 | 0.147 |
| 12 | 0.762 | 0.015 | 0.084 | 0.14 |
| 13 | 0.769 | 0.001 | 0.011 | 0.219 |
| 14 | 0.0 | 0.999 | 0.001 | 0.0 |
| 15 | 0.004 | 0.794 | 0.001 | 0.201 |
| 16 | 0.223 | 0.0 | 0.771 | 0.006 |
| 17 | 0.001 | 0.029 | 0.97 | 0.0 |
| 18 | 0.313 | 0.098 | 0.005 | 0.584 |
| 19 | 0.224 | 0.173 | 0.066 | 0.536 |
| 20 | 0.239 | 0.263 | 0.173 | 0.325 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.294 | -0.253 | 0.054 | -0.191 |
| 02 | 0.836 | -1.172 | -0.399 | -0.339 |
| 03 | 0.723 | -2.056 | -0.297 | 0.066 |
| 04 | -3.627 | 1.281 | -1.287 | -2.316 |
| 05 | -1.376 | 0.916 | -2.085 | 0.118 |
| 06 | 0.227 | -2.066 | 0.862 | -1.381 |
| 07 | -2.276 | -1.107 | 1.264 | -3.568 |
| 08 | 0.109 | 0.055 | -2.167 | 0.539 |
| 09 | -0.361 | -0.014 | -0.999 | 0.667 |
| 10 | -0.097 | -0.077 | -0.175 | 0.283 |
| 11 | 0.573 | -1.145 | 0.277 | -0.528 |
| 12 | 1.114 | -2.814 | -1.093 | -0.582 |
| 13 | 1.123 | -5.819 | -3.103 | -0.132 |
| 14 | -6.613 | 1.385 | -5.69 | -6.974 |
| 15 | -4.187 | 1.155 | -5.079 | -0.218 |
| 16 | -0.114 | -6.974 | 1.126 | -3.758 |
| 17 | -5.474 | -2.136 | 1.355 | -6.613 |
| 18 | 0.225 | -0.935 | -3.849 | 0.847 |
| 19 | -0.11 | -0.366 | -1.322 | 0.763 |
| 20 | -0.044 | 0.05 | -0.37 | 0.263 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.60546 |
| 0.0005 | 3.92531 |
| 0.0001 | 6.56921 |