| Motif | TF2LY.H12INVIVO.0.SM.D |
| Gene (human) | TGIF2LY (GeneCards) |
| Gene synonyms (human) | TGIFLY |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 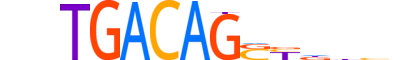 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | D |
| Motif | TF2LY.H12INVIVO.0.SM.D |
| Gene (human) | TGIF2LY (GeneCards) |
| Gene synonyms (human) | TGIFLY |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 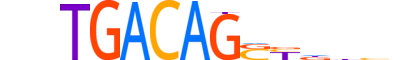 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | D |
| Motif length | 14 |
| Consensus | nnTGACAGSYddhn |
| GC content | 50.72% |
| Information content (bits; total / per base) | 13.0 / 0.929 |
| Data sources | HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 9969 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.989 | 0.983 | 0.976 | 0.965 | 0.931 | 0.916 |
| best | 0.989 | 0.984 | 0.976 | 0.965 | 0.941 | 0.924 | |
| Methyl HT-SELEX, 1 experiments | median | 0.989 | 0.984 | 0.976 | 0.965 | 0.941 | 0.924 |
| best | 0.989 | 0.984 | 0.976 | 0.965 | 0.941 | 0.924 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.989 | 0.983 | 0.976 | 0.965 | 0.921 | 0.909 |
| best | 0.989 | 0.983 | 0.976 | 0.965 | 0.921 | 0.909 | |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | TALE-type HD {3.1.4} (TFClass) |
| TF subfamily | PKNOX {3.1.4.5} (TFClass) |
| TFClass ID | TFClass: 3.1.4.5.4 |
| HGNC | HGNC:18569 |
| MGI | |
| EntrezGene (human) | GeneID:90655 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | TF2LY_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q8IUE0 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| PCM | TF2LY.H12INVIVO.0.SM.D.pcm |
| PWM | TF2LY.H12INVIVO.0.SM.D.pwm |
| PFM | TF2LY.H12INVIVO.0.SM.D.pfm |
| Alignment | TF2LY.H12INVIVO.0.SM.D.words.tsv |
| Threshold to P-value map | TF2LY.H12INVIVO.0.SM.D.thr |
| Motif in other formats | |
| JASPAR format | TF2LY.H12INVIVO.0.SM.D_jaspar_format.txt |
| MEME format | TF2LY.H12INVIVO.0.SM.D_meme_format.meme |
| Transfac format | TF2LY.H12INVIVO.0.SM.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 2959.5 | 2071.5 | 2477.5 | 2460.5 |
| 02 | 2544.75 | 1739.75 | 2193.75 | 3490.75 |
| 03 | 65.0 | 46.0 | 9.0 | 9849.0 |
| 04 | 0.0 | 0.0 | 9969.0 | 0.0 |
| 05 | 9912.0 | 0.0 | 0.0 | 57.0 |
| 06 | 0.0 | 9969.0 | 0.0 | 0.0 |
| 07 | 9931.0 | 0.0 | 38.0 | 0.0 |
| 08 | 95.0 | 99.0 | 9442.0 | 333.0 |
| 09 | 664.0 | 6165.0 | 2669.0 | 471.0 |
| 10 | 598.0 | 2042.0 | 1354.0 | 5975.0 |
| 11 | 1764.0 | 1371.0 | 4813.0 | 2021.0 |
| 12 | 2121.0 | 1647.0 | 2178.0 | 4023.0 |
| 13 | 1881.5 | 4478.5 | 1678.5 | 1930.5 |
| 14 | 3584.5 | 2044.5 | 2286.5 | 2053.5 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.297 | 0.208 | 0.249 | 0.247 |
| 02 | 0.255 | 0.175 | 0.22 | 0.35 |
| 03 | 0.007 | 0.005 | 0.001 | 0.988 |
| 04 | 0.0 | 0.0 | 1.0 | 0.0 |
| 05 | 0.994 | 0.0 | 0.0 | 0.006 |
| 06 | 0.0 | 1.0 | 0.0 | 0.0 |
| 07 | 0.996 | 0.0 | 0.004 | 0.0 |
| 08 | 0.01 | 0.01 | 0.947 | 0.033 |
| 09 | 0.067 | 0.618 | 0.268 | 0.047 |
| 10 | 0.06 | 0.205 | 0.136 | 0.599 |
| 11 | 0.177 | 0.138 | 0.483 | 0.203 |
| 12 | 0.213 | 0.165 | 0.218 | 0.404 |
| 13 | 0.189 | 0.449 | 0.168 | 0.194 |
| 14 | 0.36 | 0.205 | 0.229 | 0.206 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.172 | -0.185 | -0.006 | -0.013 |
| 02 | 0.021 | -0.359 | -0.127 | 0.337 |
| 03 | -3.613 | -3.944 | -5.397 | 1.373 |
| 04 | -6.988 | -6.988 | 1.386 | -6.988 |
| 05 | 1.38 | -6.988 | -6.988 | -3.739 |
| 06 | -6.988 | 1.386 | -6.988 | -6.988 |
| 07 | 1.382 | -6.988 | -4.125 | -6.988 |
| 08 | -3.244 | -3.204 | 1.331 | -2.007 |
| 09 | -1.32 | 0.905 | 0.068 | -1.662 |
| 10 | -1.424 | -0.199 | -0.609 | 0.874 |
| 11 | -0.345 | -0.597 | 0.658 | -0.209 |
| 12 | -0.161 | -0.414 | -0.135 | 0.478 |
| 13 | -0.281 | 0.586 | -0.395 | -0.255 |
| 14 | 0.363 | -0.198 | -0.086 | -0.193 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.59711 |
| 0.0005 | 4.07851 |
| 0.0001 | 7.436865 |