| Motif | TF2LX.H12RSNP.1.SM.D |
| Gene (human) | TGIF2LX (GeneCards) |
| Gene synonyms (human) | TGIFLX |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 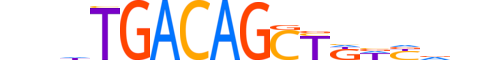 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | D |
| Motif | TF2LX.H12RSNP.1.SM.D |
| Gene (human) | TGIF2LX (GeneCards) |
| Gene synonyms (human) | TGIFLX |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 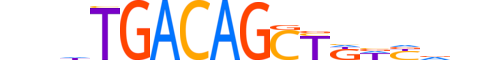 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | D |
| Motif length | 16 |
| Consensus | nndTGACAGCTKdYdn |
| GC content | 49.42% |
| Information content (bits; total / per base) | 15.781 / 0.986 |
| Data sources | HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 2234 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 6 experiments | median | 0.947 | 0.921 | 0.89 | 0.858 | 0.786 | 0.768 |
| best | 0.997 | 0.996 | 0.99 | 0.986 | 0.965 | 0.956 | |
| Methyl HT-SELEX, 2 experiments | median | 0.969 | 0.955 | 0.932 | 0.914 | 0.862 | 0.851 |
| best | 0.997 | 0.996 | 0.99 | 0.986 | 0.965 | 0.956 | |
| Non-Methyl HT-SELEX, 4 experiments | median | 0.758 | 0.74 | 0.72 | 0.702 | 0.666 | 0.655 |
| best | 0.994 | 0.992 | 0.986 | 0.98 | 0.948 | 0.938 | |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | TALE-type HD {3.1.4} (TFClass) |
| TF subfamily | PKNOX {3.1.4.5} (TFClass) |
| TFClass ID | TFClass: 3.1.4.5.3 |
| HGNC | HGNC:18570 |
| MGI | |
| EntrezGene (human) | GeneID:90316 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | TF2LX_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q8IUE1 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 4 |
| Methyl-HT-SELEX | 2 |
| PCM | TF2LX.H12RSNP.1.SM.D.pcm |
| PWM | TF2LX.H12RSNP.1.SM.D.pwm |
| PFM | TF2LX.H12RSNP.1.SM.D.pfm |
| Alignment | TF2LX.H12RSNP.1.SM.D.words.tsv |
| Threshold to P-value map | TF2LX.H12RSNP.1.SM.D.thr |
| Motif in other formats | |
| JASPAR format | TF2LX.H12RSNP.1.SM.D_jaspar_format.txt |
| MEME format | TF2LX.H12RSNP.1.SM.D_meme_format.meme |
| Transfac format | TF2LX.H12RSNP.1.SM.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 648.5 | 574.5 | 490.5 | 520.5 |
| 02 | 796.25 | 363.25 | 436.25 | 638.25 |
| 03 | 576.0 | 298.0 | 370.0 | 990.0 |
| 04 | 4.0 | 71.0 | 0.0 | 2159.0 |
| 05 | 0.0 | 0.0 | 2234.0 | 0.0 |
| 06 | 2232.0 | 0.0 | 0.0 | 2.0 |
| 07 | 0.0 | 2234.0 | 0.0 | 0.0 |
| 08 | 2234.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 5.0 | 2218.0 | 11.0 |
| 10 | 42.0 | 1840.0 | 333.0 | 19.0 |
| 11 | 40.0 | 226.0 | 183.0 | 1785.0 |
| 12 | 269.0 | 173.0 | 1484.0 | 308.0 |
| 13 | 407.0 | 211.0 | 295.0 | 1321.0 |
| 14 | 234.0 | 1418.0 | 245.0 | 337.0 |
| 15 | 1064.5 | 296.5 | 520.5 | 352.5 |
| 16 | 446.25 | 528.25 | 616.25 | 643.25 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.29 | 0.257 | 0.22 | 0.233 |
| 02 | 0.356 | 0.163 | 0.195 | 0.286 |
| 03 | 0.258 | 0.133 | 0.166 | 0.443 |
| 04 | 0.002 | 0.032 | 0.0 | 0.966 |
| 05 | 0.0 | 0.0 | 1.0 | 0.0 |
| 06 | 0.999 | 0.0 | 0.0 | 0.001 |
| 07 | 0.0 | 1.0 | 0.0 | 0.0 |
| 08 | 1.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.002 | 0.993 | 0.005 |
| 10 | 0.019 | 0.824 | 0.149 | 0.009 |
| 11 | 0.018 | 0.101 | 0.082 | 0.799 |
| 12 | 0.12 | 0.077 | 0.664 | 0.138 |
| 13 | 0.182 | 0.094 | 0.132 | 0.591 |
| 14 | 0.105 | 0.635 | 0.11 | 0.151 |
| 15 | 0.476 | 0.133 | 0.233 | 0.158 |
| 16 | 0.2 | 0.236 | 0.276 | 0.288 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.149 | 0.028 | -0.129 | -0.07 |
| 02 | 0.354 | -0.428 | -0.246 | 0.133 |
| 03 | 0.031 | -0.625 | -0.41 | 0.571 |
| 04 | -4.549 | -2.039 | -5.672 | 1.35 |
| 05 | -5.672 | -5.672 | 1.384 | -5.672 |
| 06 | 1.383 | -5.672 | -5.672 | -4.961 |
| 07 | -5.672 | 1.384 | -5.672 | -5.672 |
| 08 | 1.384 | -5.672 | -5.672 | -5.672 |
| 09 | -5.672 | -4.393 | 1.377 | -3.769 |
| 10 | -2.546 | 1.19 | -0.515 | -3.288 |
| 11 | -2.593 | -0.9 | -1.109 | 1.16 |
| 12 | -0.727 | -1.164 | 0.975 | -0.592 |
| 13 | -0.315 | -0.968 | -0.635 | 0.859 |
| 14 | -0.865 | 0.93 | -0.82 | -0.503 |
| 15 | 0.643 | -0.63 | -0.07 | -0.458 |
| 16 | -0.224 | -0.055 | 0.098 | 0.141 |
| P-value | Threshold |
|---|---|
| 0.001 | 1.10046 |
| 0.0005 | 2.63901 |
| 0.0001 | 6.01891 |