| Motif | TEAD3.H12INVIVO.1.S.B |
| Gene (human) | TEAD3 (GeneCards) |
| Gene synonyms (human) | TEAD5, TEF5 |
| Gene (mouse) | Tead3 |
| Gene synonyms (mouse) | Tcf13r2, Tef5 |
| LOGO | 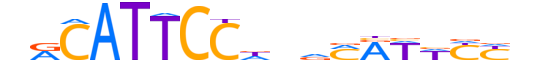 |
| LOGO (reverse complement) | 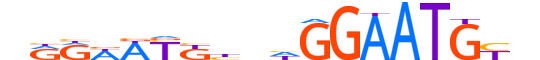 |
| Motif subtype | 1 |
| Quality | B |
| Motif | TEAD3.H12INVIVO.1.S.B |
| Gene (human) | TEAD3 (GeneCards) |
| Gene synonyms (human) | TEAD5, TEF5 |
| Gene (mouse) | Tead3 |
| Gene synonyms (mouse) | Tcf13r2, Tef5 |
| LOGO | 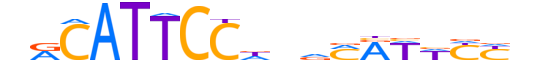 |
| LOGO (reverse complement) | 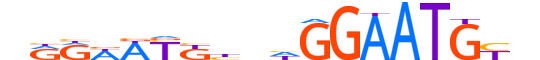 |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 18 |
| Consensus | nRCATTCCdndMRWWYYn |
| GC content | 44.94% |
| Information content (bits; total / per base) | 15.221 / 0.846 |
| Data sources | HT-SELEX |
| Aligned words | 2493 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (7) | 0.819 | 0.826 | 0.714 | 0.729 | 0.82 | 0.829 | 3.035 | 3.198 | 226.229 | 266.886 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 2 experiments | median | 0.998 | 0.998 | 0.973 | 0.971 | 0.891 | 0.896 |
| best | 1.0 | 1.0 | 1.0 | 0.999 | 0.978 | 0.978 | |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | TEA domain factors {3.6} (TFClass) |
| TF family | TEF1-related {3.6.1} (TFClass) |
| TF subfamily | {3.6.1.0} (TFClass) |
| TFClass ID | TFClass: 3.6.1.0.4 |
| HGNC | HGNC:11716 |
| MGI | MGI:109241 |
| EntrezGene (human) | GeneID:7005 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:21678 (SSTAR profile) |
| UniProt ID (human) | TEAD3_HUMAN |
| UniProt ID (mouse) | TEAD3_MOUSE |
| UniProt AC (human) | Q99594 (TFClass) |
| UniProt AC (mouse) | P70210 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 0 |
| PCM | TEAD3.H12INVIVO.1.S.B.pcm |
| PWM | TEAD3.H12INVIVO.1.S.B.pwm |
| PFM | TEAD3.H12INVIVO.1.S.B.pfm |
| Alignment | TEAD3.H12INVIVO.1.S.B.words.tsv |
| Threshold to P-value map | TEAD3.H12INVIVO.1.S.B.thr |
| Motif in other formats | |
| JASPAR format | TEAD3.H12INVIVO.1.S.B_jaspar_format.txt |
| MEME format | TEAD3.H12INVIVO.1.S.B_meme_format.meme |
| Transfac format | TEAD3.H12INVIVO.1.S.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 468.0 | 815.0 | 438.0 | 772.0 |
| 02 | 1220.0 | 85.0 | 1101.0 | 87.0 |
| 03 | 274.0 | 2178.0 | 35.0 | 6.0 |
| 04 | 2467.0 | 5.0 | 19.0 | 2.0 |
| 05 | 6.0 | 3.0 | 3.0 | 2481.0 |
| 06 | 160.0 | 5.0 | 20.0 | 2308.0 |
| 07 | 4.0 | 2439.0 | 25.0 | 25.0 |
| 08 | 14.0 | 2204.0 | 25.0 | 250.0 |
| 09 | 1006.0 | 144.0 | 359.0 | 984.0 |
| 10 | 633.0 | 626.0 | 553.0 | 681.0 |
| 11 | 928.0 | 258.0 | 999.0 | 308.0 |
| 12 | 528.0 | 1607.0 | 244.0 | 114.0 |
| 13 | 1879.0 | 212.0 | 220.0 | 182.0 |
| 14 | 258.0 | 233.0 | 153.0 | 1849.0 |
| 15 | 429.0 | 238.0 | 260.0 | 1566.0 |
| 16 | 243.0 | 1791.0 | 178.0 | 281.0 |
| 17 | 254.25 | 1634.25 | 158.25 | 446.25 |
| 18 | 847.25 | 449.25 | 450.25 | 746.25 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.188 | 0.327 | 0.176 | 0.31 |
| 02 | 0.489 | 0.034 | 0.442 | 0.035 |
| 03 | 0.11 | 0.874 | 0.014 | 0.002 |
| 04 | 0.99 | 0.002 | 0.008 | 0.001 |
| 05 | 0.002 | 0.001 | 0.001 | 0.995 |
| 06 | 0.064 | 0.002 | 0.008 | 0.926 |
| 07 | 0.002 | 0.978 | 0.01 | 0.01 |
| 08 | 0.006 | 0.884 | 0.01 | 0.1 |
| 09 | 0.404 | 0.058 | 0.144 | 0.395 |
| 10 | 0.254 | 0.251 | 0.222 | 0.273 |
| 11 | 0.372 | 0.103 | 0.401 | 0.124 |
| 12 | 0.212 | 0.645 | 0.098 | 0.046 |
| 13 | 0.754 | 0.085 | 0.088 | 0.073 |
| 14 | 0.103 | 0.093 | 0.061 | 0.742 |
| 15 | 0.172 | 0.095 | 0.104 | 0.628 |
| 16 | 0.097 | 0.718 | 0.071 | 0.113 |
| 17 | 0.102 | 0.656 | 0.063 | 0.179 |
| 18 | 0.34 | 0.18 | 0.181 | 0.299 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.285 | 0.268 | -0.351 | 0.213 |
| 02 | 0.67 | -1.973 | 0.568 | -1.95 |
| 03 | -0.818 | 1.249 | -2.828 | -4.364 |
| 04 | 1.373 | -4.499 | -3.396 | -5.063 |
| 05 | -4.364 | -4.838 | -4.838 | 1.379 |
| 06 | -1.351 | -4.499 | -3.349 | 1.307 |
| 07 | -4.654 | 1.362 | -3.144 | -3.144 |
| 08 | -3.668 | 1.261 | -3.144 | -0.909 |
| 09 | 0.478 | -1.455 | -0.549 | 0.456 |
| 10 | 0.015 | 0.004 | -0.119 | 0.088 |
| 11 | 0.397 | -0.878 | 0.471 | -0.702 |
| 12 | -0.165 | 0.945 | -0.933 | -1.685 |
| 13 | 1.101 | -1.072 | -1.036 | -1.223 |
| 14 | -0.878 | -0.979 | -1.395 | 1.085 |
| 15 | -0.372 | -0.958 | -0.87 | 0.919 |
| 16 | -0.937 | 1.054 | -1.245 | -0.793 |
| 17 | -0.892 | 0.962 | -1.362 | -0.333 |
| 18 | 0.306 | -0.326 | -0.324 | 0.18 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.73571 |
| 0.0005 | 3.97046 |
| 0.0001 | 6.52166 |