| Motif | TEAD3.H12INVITRO.1.S.B |
| Gene (human) | TEAD3 (GeneCards) |
| Gene synonyms (human) | TEAD5, TEF5 |
| Gene (mouse) | Tead3 |
| Gene synonyms (mouse) | Tcf13r2, Tef5 |
| LOGO | 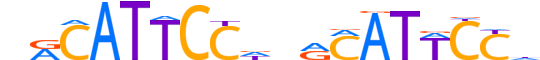 |
| LOGO (reverse complement) | 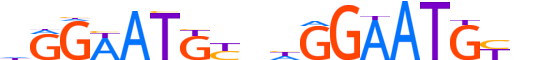 |
| Motif subtype | 1 |
| Quality | B |
| Motif | TEAD3.H12INVITRO.1.S.B |
| Gene (human) | TEAD3 (GeneCards) |
| Gene synonyms (human) | TEAD5, TEF5 |
| Gene (mouse) | Tead3 |
| Gene synonyms (mouse) | Tcf13r2, Tef5 |
| LOGO |  |
| LOGO (reverse complement) | 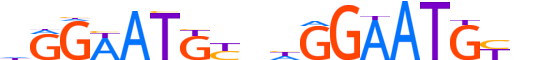 |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 18 |
| Consensus | nRCATTCCdnRCATWCCd |
| GC content | 44.19% |
| Information content (bits; total / per base) | 19.592 / 1.088 |
| Data sources | HT-SELEX |
| Aligned words | 7183 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (7) | 0.723 | 0.735 | 0.624 | 0.643 | 0.731 | 0.746 | 2.745 | 2.941 | 135.292 | 165.018 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 2 experiments | median | 0.999 | 0.999 | 0.974 | 0.972 | 0.888 | 0.895 |
| best | 1.0 | 1.0 | 1.0 | 1.0 | 0.977 | 0.977 | |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | TEA domain factors {3.6} (TFClass) |
| TF family | TEF1-related {3.6.1} (TFClass) |
| TF subfamily | {3.6.1.0} (TFClass) |
| TFClass ID | TFClass: 3.6.1.0.4 |
| HGNC | HGNC:11716 |
| MGI | MGI:109241 |
| EntrezGene (human) | GeneID:7005 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:21678 (SSTAR profile) |
| UniProt ID (human) | TEAD3_HUMAN |
| UniProt ID (mouse) | TEAD3_MOUSE |
| UniProt AC (human) | Q99594 (TFClass) |
| UniProt AC (mouse) | P70210 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 0 |
| PCM | TEAD3.H12INVITRO.1.S.B.pcm |
| PWM | TEAD3.H12INVITRO.1.S.B.pwm |
| PFM | TEAD3.H12INVITRO.1.S.B.pfm |
| Alignment | TEAD3.H12INVITRO.1.S.B.words.tsv |
| Threshold to P-value map | TEAD3.H12INVITRO.1.S.B.thr |
| Motif in other formats | |
| JASPAR format | TEAD3.H12INVITRO.1.S.B_jaspar_format.txt |
| MEME format | TEAD3.H12INVITRO.1.S.B_meme_format.meme |
| Transfac format | TEAD3.H12INVITRO.1.S.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1460.0 | 2467.0 | 1202.0 | 2054.0 |
| 02 | 3599.5 | 164.5 | 3205.5 | 213.5 |
| 03 | 849.0 | 6221.0 | 97.0 | 16.0 |
| 04 | 7132.0 | 11.0 | 39.0 | 1.0 |
| 05 | 2.0 | 4.0 | 4.0 | 7173.0 |
| 06 | 608.0 | 7.0 | 93.0 | 6475.0 |
| 07 | 33.0 | 7059.0 | 53.0 | 38.0 |
| 08 | 50.0 | 6163.0 | 48.0 | 922.0 |
| 09 | 2591.0 | 418.0 | 938.0 | 3236.0 |
| 10 | 1901.0 | 2000.0 | 1609.0 | 1673.0 |
| 11 | 3126.0 | 214.0 | 3454.0 | 389.0 |
| 12 | 1347.0 | 5557.0 | 228.0 | 51.0 |
| 13 | 6827.0 | 90.0 | 223.0 | 43.0 |
| 14 | 108.0 | 132.0 | 55.0 | 6888.0 |
| 15 | 1272.0 | 94.0 | 381.0 | 5436.0 |
| 16 | 196.0 | 6545.0 | 185.0 | 257.0 |
| 17 | 257.25 | 5850.25 | 169.25 | 906.25 |
| 18 | 2708.5 | 831.5 | 1319.5 | 2323.5 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.203 | 0.343 | 0.167 | 0.286 |
| 02 | 0.501 | 0.023 | 0.446 | 0.03 |
| 03 | 0.118 | 0.866 | 0.014 | 0.002 |
| 04 | 0.993 | 0.002 | 0.005 | 0.0 |
| 05 | 0.0 | 0.001 | 0.001 | 0.999 |
| 06 | 0.085 | 0.001 | 0.013 | 0.901 |
| 07 | 0.005 | 0.983 | 0.007 | 0.005 |
| 08 | 0.007 | 0.858 | 0.007 | 0.128 |
| 09 | 0.361 | 0.058 | 0.131 | 0.451 |
| 10 | 0.265 | 0.278 | 0.224 | 0.233 |
| 11 | 0.435 | 0.03 | 0.481 | 0.054 |
| 12 | 0.188 | 0.774 | 0.032 | 0.007 |
| 13 | 0.95 | 0.013 | 0.031 | 0.006 |
| 14 | 0.015 | 0.018 | 0.008 | 0.959 |
| 15 | 0.177 | 0.013 | 0.053 | 0.757 |
| 16 | 0.027 | 0.911 | 0.026 | 0.036 |
| 17 | 0.036 | 0.814 | 0.024 | 0.126 |
| 18 | 0.377 | 0.116 | 0.184 | 0.323 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.207 | 0.317 | -0.401 | 0.134 |
| 02 | 0.695 | -2.378 | 0.579 | -2.12 |
| 03 | -0.748 | 1.242 | -2.897 | -4.592 |
| 04 | 1.378 | -4.913 | -3.775 | -6.325 |
| 05 | -6.055 | -5.667 | -5.667 | 1.384 |
| 06 | -1.081 | -5.273 | -2.938 | 1.282 |
| 07 | -3.933 | 1.368 | -3.483 | -3.8 |
| 08 | -3.539 | 1.232 | -3.578 | -0.665 |
| 09 | 0.366 | -1.454 | -0.648 | 0.588 |
| 10 | 0.057 | 0.108 | -0.11 | -0.071 |
| 11 | 0.554 | -2.118 | 0.654 | -1.525 |
| 12 | -0.287 | 1.129 | -2.055 | -3.52 |
| 13 | 1.335 | -2.97 | -2.077 | -3.683 |
| 14 | -2.792 | -2.595 | -3.448 | 1.343 |
| 15 | -0.344 | -2.928 | -1.546 | 1.107 |
| 16 | -2.205 | 1.292 | -2.262 | -1.937 |
| 17 | -1.936 | 1.18 | -2.35 | -0.683 |
| 18 | 0.411 | -0.769 | -0.308 | 0.257 |
| P-value | Threshold |
|---|---|
| 0.001 | -0.26569 |
| 0.0005 | 1.27781 |
| 0.0001 | 4.51586 |