| Motif | TEAD3.H12CORE.1.S.B |
| Gene (human) | TEAD3 (GeneCards) |
| Gene synonyms (human) | TEAD5, TEF5 |
| Gene (mouse) | Tead3 |
| Gene synonyms (mouse) | Tcf13r2, Tef5 |
| LOGO | 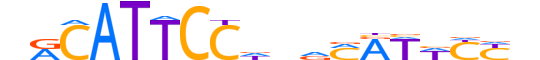 |
| LOGO (reverse complement) | 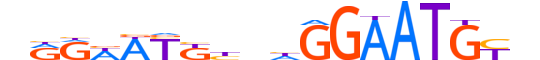 |
| Motif subtype | 1 |
| Quality | B |
| Motif | TEAD3.H12CORE.1.S.B |
| Gene (human) | TEAD3 (GeneCards) |
| Gene synonyms (human) | TEAD5, TEF5 |
| Gene (mouse) | Tead3 |
| Gene synonyms (mouse) | Tcf13r2, Tef5 |
| LOGO | 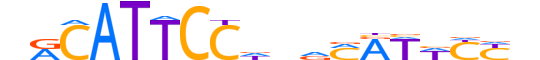 |
| LOGO (reverse complement) | 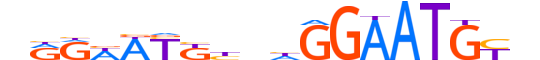 |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 18 |
| Consensus | nRCATTCCdndMRWWYYn |
| GC content | 44.72% |
| Information content (bits; total / per base) | 15.7 / 0.872 |
| Data sources | HT-SELEX |
| Aligned words | 9831 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (7) | 0.806 | 0.813 | 0.699 | 0.713 | 0.811 | 0.819 | 3.02 | 3.18 | 212.796 | 243.347 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 2 experiments | median | 0.999 | 0.998 | 0.974 | 0.972 | 0.893 | 0.898 |
| best | 1.0 | 1.0 | 1.0 | 1.0 | 0.979 | 0.979 | |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | TEA domain factors {3.6} (TFClass) |
| TF family | TEF1-related {3.6.1} (TFClass) |
| TF subfamily | {3.6.1.0} (TFClass) |
| TFClass ID | TFClass: 3.6.1.0.4 |
| HGNC | HGNC:11716 |
| MGI | MGI:109241 |
| EntrezGene (human) | GeneID:7005 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:21678 (SSTAR profile) |
| UniProt ID (human) | TEAD3_HUMAN |
| UniProt ID (mouse) | TEAD3_MOUSE |
| UniProt AC (human) | Q99594 (TFClass) |
| UniProt AC (mouse) | P70210 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 0 |
| PCM | TEAD3.H12CORE.1.S.B.pcm |
| PWM | TEAD3.H12CORE.1.S.B.pwm |
| PFM | TEAD3.H12CORE.1.S.B.pfm |
| Alignment | TEAD3.H12CORE.1.S.B.words.tsv |
| Threshold to P-value map | TEAD3.H12CORE.1.S.B.thr |
| Motif in other formats | |
| JASPAR format | TEAD3.H12CORE.1.S.B_jaspar_format.txt |
| MEME format | TEAD3.H12CORE.1.S.B_meme_format.meme |
| Transfac format | TEAD3.H12CORE.1.S.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1948.5 | 3354.5 | 1674.5 | 2853.5 |
| 02 | 4869.5 | 206.5 | 4478.5 | 276.5 |
| 03 | 1174.0 | 8508.0 | 121.0 | 28.0 |
| 04 | 9746.0 | 19.0 | 63.0 | 3.0 |
| 05 | 6.0 | 9.0 | 5.0 | 9811.0 |
| 06 | 777.0 | 12.0 | 148.0 | 8894.0 |
| 07 | 46.0 | 9639.0 | 80.0 | 66.0 |
| 08 | 72.0 | 8504.0 | 83.0 | 1172.0 |
| 09 | 3605.0 | 560.0 | 1398.0 | 4268.0 |
| 10 | 2561.0 | 2713.0 | 2181.0 | 2376.0 |
| 11 | 3950.0 | 767.0 | 4211.0 | 903.0 |
| 12 | 2081.0 | 6431.0 | 912.0 | 407.0 |
| 13 | 7688.0 | 722.0 | 818.0 | 603.0 |
| 14 | 858.0 | 833.0 | 504.0 | 7636.0 |
| 15 | 2044.0 | 811.0 | 921.0 | 6055.0 |
| 16 | 920.0 | 7335.0 | 558.0 | 1018.0 |
| 17 | 827.25 | 6581.25 | 569.25 | 1853.25 |
| 18 | 3318.25 | 1650.25 | 1754.25 | 3108.25 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.198 | 0.341 | 0.17 | 0.29 |
| 02 | 0.495 | 0.021 | 0.456 | 0.028 |
| 03 | 0.119 | 0.865 | 0.012 | 0.003 |
| 04 | 0.991 | 0.002 | 0.006 | 0.0 |
| 05 | 0.001 | 0.001 | 0.001 | 0.998 |
| 06 | 0.079 | 0.001 | 0.015 | 0.905 |
| 07 | 0.005 | 0.98 | 0.008 | 0.007 |
| 08 | 0.007 | 0.865 | 0.008 | 0.119 |
| 09 | 0.367 | 0.057 | 0.142 | 0.434 |
| 10 | 0.261 | 0.276 | 0.222 | 0.242 |
| 11 | 0.402 | 0.078 | 0.428 | 0.092 |
| 12 | 0.212 | 0.654 | 0.093 | 0.041 |
| 13 | 0.782 | 0.073 | 0.083 | 0.061 |
| 14 | 0.087 | 0.085 | 0.051 | 0.777 |
| 15 | 0.208 | 0.082 | 0.094 | 0.616 |
| 16 | 0.094 | 0.746 | 0.057 | 0.104 |
| 17 | 0.084 | 0.669 | 0.058 | 0.189 |
| 18 | 0.338 | 0.168 | 0.178 | 0.316 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.232 | 0.311 | -0.383 | 0.149 |
| 02 | 0.683 | -2.467 | 0.6 | -2.177 |
| 03 | -0.738 | 1.241 | -2.993 | -4.397 |
| 04 | 1.377 | -4.749 | -3.629 | -6.141 |
| 05 | -5.692 | -5.383 | -5.82 | 1.384 |
| 06 | -1.15 | -5.148 | -2.795 | 1.285 |
| 07 | -3.931 | 1.366 | -3.398 | -3.584 |
| 08 | -3.5 | 1.241 | -3.362 | -0.74 |
| 09 | 0.383 | -1.476 | -0.563 | 0.552 |
| 10 | 0.041 | 0.099 | -0.119 | -0.034 |
| 11 | 0.474 | -1.162 | 0.538 | -1.0 |
| 12 | -0.166 | 0.961 | -0.99 | -1.793 |
| 13 | 1.14 | -1.223 | -1.098 | -1.402 |
| 14 | -1.051 | -1.08 | -1.581 | 1.133 |
| 15 | -0.184 | -1.107 | -0.98 | 0.901 |
| 16 | -0.981 | 1.093 | -1.479 | -0.88 |
| 17 | -1.087 | 0.984 | -1.46 | -0.282 |
| 18 | 0.3 | -0.398 | -0.337 | 0.235 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.38371 |
| 0.0005 | 3.68176 |
| 0.0001 | 6.34506 |