| Motif | TCF21.H12CORE.0.PS.A |
| Gene (human) | TCF21 (GeneCards) |
| Gene synonyms (human) | BHLHA23, POD1 |
| Gene (mouse) | Tcf21 |
| Gene synonyms (mouse) | Pod1 |
| LOGO | 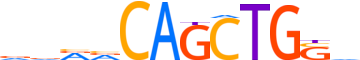 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | A |
| Motif | TCF21.H12CORE.0.PS.A |
| Gene (human) | TCF21 (GeneCards) |
| Gene synonyms (human) | BHLHA23, POD1 |
| Gene (mouse) | Tcf21 |
| Gene synonyms (mouse) | Pod1 |
| LOGO | 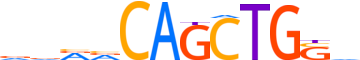 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 12 |
| Consensus | dvvvCAGCTGSv |
| GC content | 57.32% |
| Information content (bits; total / per base) | 12.263 / 1.022 |
| Data sources | ChIP-Seq + HT-SELEX |
| Aligned words | 1000 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 3 (9) | 0.928 | 0.934 | 0.857 | 0.864 | 0.896 | 0.902 | 3.281 | 3.356 | 557.0 | 712.854 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 5 experiments | median | 0.872 | 0.826 | 0.777 | 0.741 | 0.67 | 0.662 |
| best | 0.924 | 0.891 | 0.872 | 0.831 | 0.739 | 0.732 | |
| Methyl HT-SELEX, 1 experiments | median | 0.924 | 0.891 | 0.872 | 0.831 | 0.739 | 0.732 |
| best | 0.924 | 0.891 | 0.872 | 0.831 | 0.739 | 0.732 | |
| Non-Methyl HT-SELEX, 4 experiments | median | 0.793 | 0.753 | 0.697 | 0.674 | 0.619 | 0.615 |
| best | 0.899 | 0.861 | 0.836 | 0.793 | 0.72 | 0.706 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.708 | 0.135 | 0.762 | 0.463 |
| TF superclass | Basic domains {1} (TFClass) |
| TF class | Basic helix-loop-helix factors (bHLH) {1.2} (TFClass) |
| TF family | Tal-related {1.2.3} (TFClass) |
| TF subfamily | MESP {1.2.3.3} (TFClass) |
| TFClass ID | TFClass: 1.2.3.3.5 |
| HGNC | HGNC:11632 |
| MGI | MGI:1202715 |
| EntrezGene (human) | GeneID:6943 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:21412 (SSTAR profile) |
| UniProt ID (human) | TCF21_HUMAN |
| UniProt ID (mouse) | TCF21_MOUSE |
| UniProt AC (human) | O43680 (TFClass) |
| UniProt AC (mouse) | O35437 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 3 human, 0 mouse |
| HT-SELEX | 4 |
| Methyl-HT-SELEX | 1 |
| PCM | TCF21.H12CORE.0.PS.A.pcm |
| PWM | TCF21.H12CORE.0.PS.A.pwm |
| PFM | TCF21.H12CORE.0.PS.A.pfm |
| Alignment | TCF21.H12CORE.0.PS.A.words.tsv |
| Threshold to P-value map | TCF21.H12CORE.0.PS.A.thr |
| Motif in other formats | |
| JASPAR format | TCF21.H12CORE.0.PS.A_jaspar_format.txt |
| MEME format | TCF21.H12CORE.0.PS.A_meme_format.meme |
| Transfac format | TCF21.H12CORE.0.PS.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 372.0 | 126.0 | 355.0 | 147.0 |
| 02 | 381.0 | 191.0 | 299.0 | 129.0 |
| 03 | 557.0 | 197.0 | 200.0 | 46.0 |
| 04 | 516.0 | 200.0 | 227.0 | 57.0 |
| 05 | 1.0 | 998.0 | 1.0 | 0.0 |
| 06 | 995.0 | 0.0 | 2.0 | 3.0 |
| 07 | 24.0 | 13.0 | 878.0 | 85.0 |
| 08 | 64.0 | 920.0 | 10.0 | 6.0 |
| 09 | 8.0 | 3.0 | 1.0 | 988.0 |
| 10 | 3.0 | 1.0 | 983.0 | 13.0 |
| 11 | 8.0 | 192.0 | 642.0 | 158.0 |
| 12 | 401.0 | 192.0 | 247.0 | 160.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.372 | 0.126 | 0.355 | 0.147 |
| 02 | 0.381 | 0.191 | 0.299 | 0.129 |
| 03 | 0.557 | 0.197 | 0.2 | 0.046 |
| 04 | 0.516 | 0.2 | 0.227 | 0.057 |
| 05 | 0.001 | 0.998 | 0.001 | 0.0 |
| 06 | 0.995 | 0.0 | 0.002 | 0.003 |
| 07 | 0.024 | 0.013 | 0.878 | 0.085 |
| 08 | 0.064 | 0.92 | 0.01 | 0.006 |
| 09 | 0.008 | 0.003 | 0.001 | 0.988 |
| 10 | 0.003 | 0.001 | 0.983 | 0.013 |
| 11 | 0.008 | 0.192 | 0.642 | 0.158 |
| 12 | 0.401 | 0.192 | 0.247 | 0.16 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.395 | -0.678 | 0.349 | -0.526 |
| 02 | 0.419 | -0.267 | 0.178 | -0.655 |
| 03 | 0.797 | -0.236 | -0.221 | -1.663 |
| 04 | 0.721 | -0.221 | -0.096 | -1.455 |
| 05 | -4.525 | 1.379 | -4.525 | -4.982 |
| 06 | 1.376 | -4.982 | -4.213 | -3.975 |
| 07 | -2.281 | -2.839 | 1.251 | -1.066 |
| 08 | -1.343 | 1.298 | -3.066 | -3.484 |
| 09 | -3.253 | -3.975 | -4.525 | 1.369 |
| 10 | -3.975 | -4.525 | 1.364 | -2.839 |
| 11 | -3.253 | -0.262 | 0.939 | -0.455 |
| 12 | 0.47 | -0.262 | -0.012 | -0.442 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.80826 |
| 0.0005 | 4.941985 |
| 0.0001 | 7.439305 |