| Motif | TBXT.H12INVIVO.0.PS.A |
| Gene (human) | TBXT (GeneCards) |
| Gene synonyms (human) | T |
| Gene (mouse) | Tbxt |
| Gene synonyms (mouse) | T |
| LOGO |  |
| LOGO (reverse complement) | 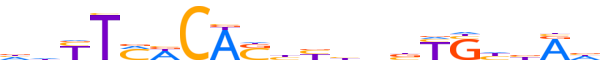 |
| Motif subtype | 0 |
| Quality | A |
| Motif | TBXT.H12INVIVO.0.PS.A |
| Gene (human) | TBXT (GeneCards) |
| Gene synonyms (human) | T |
| Gene (mouse) | Tbxt |
| Gene synonyms (mouse) | T |
| LOGO |  |
| LOGO (reverse complement) | 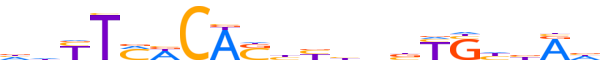 |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 20 |
| Consensus | dYvdSRvndvdSTGWKARdd |
| GC content | 45.3% |
| Information content (bits; total / per base) | 11.588 / 0.579 |
| Data sources | ChIP-Seq + HT-SELEX |
| Aligned words | 845 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 2 (14) | 0.821 | 0.853 | 0.746 | 0.768 | 0.801 | 0.843 | 3.045 | 3.215 | 104.051 | 143.921 |
| Mouse | 6 (36) | 0.795 | 0.972 | 0.775 | 0.955 | 0.784 | 0.959 | 3.999 | 6.19 | 195.674 | 473.745 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 6 experiments | median | 0.988 | 0.982 | 0.97 | 0.958 | 0.927 | 0.911 |
| best | 0.999 | 0.998 | 0.995 | 0.992 | 0.978 | 0.972 | |
| Methyl HT-SELEX, 2 experiments | median | 0.988 | 0.981 | 0.97 | 0.958 | 0.927 | 0.911 |
| best | 0.996 | 0.994 | 0.987 | 0.982 | 0.959 | 0.948 | |
| Non-Methyl HT-SELEX, 4 experiments | median | 0.983 | 0.982 | 0.873 | 0.883 | 0.793 | 0.808 |
| best | 0.999 | 0.998 | 0.995 | 0.992 | 0.978 | 0.972 | |
| TF superclass | Immunoglobulin fold {6} (TFClass) |
| TF class | T-Box factors {6.5} (TFClass) |
| TF family | Brachyury-related {6.5.1} (TFClass) |
| TF subfamily | {6.5.1.0} (TFClass) |
| TFClass ID | TFClass: 6.5.1.0.1 |
| HGNC | HGNC:11515 |
| MGI | MGI:98472 |
| EntrezGene (human) | GeneID:6862 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:20997 (SSTAR profile) |
| UniProt ID (human) | TBXT_HUMAN |
| UniProt ID (mouse) | TBXT_MOUSE |
| UniProt AC (human) | O15178 (TFClass) |
| UniProt AC (mouse) | P20293 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 6 mouse |
| HT-SELEX | 4 |
| Methyl-HT-SELEX | 2 |
| PCM | TBXT.H12INVIVO.0.PS.A.pcm |
| PWM | TBXT.H12INVIVO.0.PS.A.pwm |
| PFM | TBXT.H12INVIVO.0.PS.A.pfm |
| Alignment | TBXT.H12INVIVO.0.PS.A.words.tsv |
| Threshold to P-value map | TBXT.H12INVIVO.0.PS.A.thr |
| Motif in other formats | |
| JASPAR format | TBXT.H12INVIVO.0.PS.A_jaspar_format.txt |
| MEME format | TBXT.H12INVIVO.0.PS.A_meme_format.meme |
| Transfac format | TBXT.H12INVIVO.0.PS.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 146.0 | 117.0 | 131.0 | 451.0 |
| 02 | 40.0 | 95.0 | 58.0 | 652.0 |
| 03 | 395.0 | 159.0 | 205.0 | 86.0 |
| 04 | 285.0 | 59.0 | 385.0 | 116.0 |
| 05 | 50.0 | 660.0 | 86.0 | 49.0 |
| 06 | 625.0 | 36.0 | 120.0 | 64.0 |
| 07 | 131.0 | 320.0 | 332.0 | 62.0 |
| 08 | 151.0 | 206.0 | 231.0 | 257.0 |
| 09 | 355.0 | 106.0 | 229.0 | 155.0 |
| 10 | 423.0 | 77.0 | 281.0 | 64.0 |
| 11 | 246.0 | 109.0 | 376.0 | 114.0 |
| 12 | 60.0 | 179.0 | 555.0 | 51.0 |
| 13 | 61.0 | 42.0 | 26.0 | 716.0 |
| 14 | 7.0 | 6.0 | 827.0 | 5.0 |
| 15 | 126.0 | 124.0 | 60.0 | 535.0 |
| 16 | 32.0 | 113.0 | 536.0 | 164.0 |
| 17 | 793.0 | 8.0 | 37.0 | 7.0 |
| 18 | 586.0 | 56.0 | 150.0 | 53.0 |
| 19 | 335.0 | 96.0 | 182.0 | 232.0 |
| 20 | 138.0 | 130.0 | 150.0 | 427.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.173 | 0.138 | 0.155 | 0.534 |
| 02 | 0.047 | 0.112 | 0.069 | 0.772 |
| 03 | 0.467 | 0.188 | 0.243 | 0.102 |
| 04 | 0.337 | 0.07 | 0.456 | 0.137 |
| 05 | 0.059 | 0.781 | 0.102 | 0.058 |
| 06 | 0.74 | 0.043 | 0.142 | 0.076 |
| 07 | 0.155 | 0.379 | 0.393 | 0.073 |
| 08 | 0.179 | 0.244 | 0.273 | 0.304 |
| 09 | 0.42 | 0.125 | 0.271 | 0.183 |
| 10 | 0.501 | 0.091 | 0.333 | 0.076 |
| 11 | 0.291 | 0.129 | 0.445 | 0.135 |
| 12 | 0.071 | 0.212 | 0.657 | 0.06 |
| 13 | 0.072 | 0.05 | 0.031 | 0.847 |
| 14 | 0.008 | 0.007 | 0.979 | 0.006 |
| 15 | 0.149 | 0.147 | 0.071 | 0.633 |
| 16 | 0.038 | 0.134 | 0.634 | 0.194 |
| 17 | 0.938 | 0.009 | 0.044 | 0.008 |
| 18 | 0.693 | 0.066 | 0.178 | 0.063 |
| 19 | 0.396 | 0.114 | 0.215 | 0.275 |
| 20 | 0.163 | 0.154 | 0.178 | 0.505 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.366 | -0.585 | -0.473 | 0.754 |
| 02 | -1.631 | -0.79 | -1.272 | 1.122 |
| 03 | 0.622 | -0.282 | -0.03 | -0.887 |
| 04 | 0.297 | -1.255 | 0.597 | -0.593 |
| 05 | -1.416 | 1.134 | -0.887 | -1.435 |
| 06 | 1.079 | -1.732 | -0.56 | -1.176 |
| 07 | -0.473 | 0.413 | 0.449 | -1.207 |
| 08 | -0.333 | -0.025 | 0.089 | 0.195 |
| 09 | 0.516 | -0.682 | 0.08 | -0.307 |
| 10 | 0.69 | -0.996 | 0.283 | -1.176 |
| 11 | 0.151 | -0.654 | 0.573 | -0.61 |
| 12 | -1.239 | -0.164 | 0.961 | -1.397 |
| 13 | -1.223 | -1.584 | -2.04 | 1.215 |
| 14 | -3.199 | -3.322 | 1.359 | -3.461 |
| 15 | -0.511 | -0.527 | -1.239 | 0.924 |
| 16 | -1.844 | -0.619 | 0.926 | -0.251 |
| 17 | 1.317 | -3.09 | -1.706 | -3.199 |
| 18 | 1.015 | -1.306 | -0.339 | -1.359 |
| 19 | 0.458 | -0.779 | -0.148 | 0.093 |
| 20 | -0.422 | -0.481 | -0.339 | 0.7 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.38036 |
| 0.0005 | 5.25446 |
| 0.0001 | 7.08191 |