| Motif | STAT1.H12RSNP.0.P.B |
| Gene (human) | STAT1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Stat1 |
| Gene synonyms (mouse) | |
| LOGO | 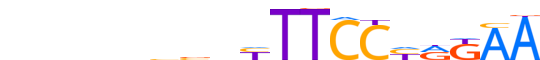 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | STAT1.H12RSNP.0.P.B |
| Gene (human) | STAT1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Stat1 |
| Gene synonyms (mouse) | |
| LOGO | 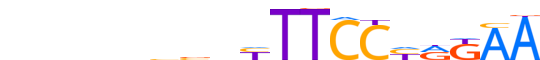 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 18 |
| Consensus | nnnnnbbnYTTCCYRKAA |
| GC content | 45.84% |
| Information content (bits; total / per base) | 11.792 / 0.655 |
| Data sources | ChIP-Seq |
| Aligned words | 1017 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 10 (65) | 0.878 | 0.974 | 0.813 | 0.954 | 0.87 | 0.967 | 3.623 | 5.701 | 182.77 | 498.745 |
| Mouse | 38 (214) | 0.819 | 0.932 | 0.729 | 0.852 | 0.813 | 0.924 | 2.904 | 3.853 | 105.325 | 355.276 |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 1.492 | 1.478 | 0.064 | 0.029 |
| TF superclass | Immunoglobulin fold {6} (TFClass) |
| TF class | STAT domain factors {6.2} (TFClass) |
| TF family | STAT {6.2.1} (TFClass) |
| TF subfamily | {6.2.1.0} (TFClass) |
| TFClass ID | TFClass: 6.2.1.0.1 |
| HGNC | HGNC:11362 |
| MGI | MGI:103063 |
| EntrezGene (human) | GeneID:6772 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | STAT1_HUMAN |
| UniProt ID (mouse) | STAT1_MOUSE |
| UniProt AC (human) | P42224 (TFClass) |
| UniProt AC (mouse) | P42225 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 10 human, 38 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | STAT1.H12RSNP.0.P.B.pcm |
| PWM | STAT1.H12RSNP.0.P.B.pwm |
| PFM | STAT1.H12RSNP.0.P.B.pfm |
| Alignment | STAT1.H12RSNP.0.P.B.words.tsv |
| Threshold to P-value map | STAT1.H12RSNP.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | STAT1.H12RSNP.0.P.B_jaspar_format.txt |
| MEME format | STAT1.H12RSNP.0.P.B_meme_format.meme |
| Transfac format | STAT1.H12RSNP.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 244.0 | 327.0 | 163.0 | 283.0 |
| 02 | 239.0 | 379.0 | 192.0 | 207.0 |
| 03 | 260.0 | 280.0 | 160.0 | 317.0 |
| 04 | 243.0 | 270.0 | 222.0 | 282.0 |
| 05 | 202.0 | 322.0 | 233.0 | 260.0 |
| 06 | 142.0 | 360.0 | 187.0 | 328.0 |
| 07 | 124.0 | 452.0 | 259.0 | 182.0 |
| 08 | 377.0 | 264.0 | 195.0 | 181.0 |
| 09 | 103.0 | 308.0 | 44.0 | 562.0 |
| 10 | 11.0 | 15.0 | 5.0 | 986.0 |
| 11 | 6.0 | 3.0 | 27.0 | 981.0 |
| 12 | 73.0 | 920.0 | 5.0 | 19.0 |
| 13 | 13.0 | 876.0 | 3.0 | 125.0 |
| 14 | 148.0 | 421.0 | 18.0 | 430.0 |
| 15 | 379.0 | 36.0 | 530.0 | 72.0 |
| 16 | 53.0 | 31.0 | 707.0 | 226.0 |
| 17 | 861.0 | 127.0 | 11.0 | 18.0 |
| 18 | 946.0 | 12.0 | 28.0 | 31.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.24 | 0.322 | 0.16 | 0.278 |
| 02 | 0.235 | 0.373 | 0.189 | 0.204 |
| 03 | 0.256 | 0.275 | 0.157 | 0.312 |
| 04 | 0.239 | 0.265 | 0.218 | 0.277 |
| 05 | 0.199 | 0.317 | 0.229 | 0.256 |
| 06 | 0.14 | 0.354 | 0.184 | 0.323 |
| 07 | 0.122 | 0.444 | 0.255 | 0.179 |
| 08 | 0.371 | 0.26 | 0.192 | 0.178 |
| 09 | 0.101 | 0.303 | 0.043 | 0.553 |
| 10 | 0.011 | 0.015 | 0.005 | 0.97 |
| 11 | 0.006 | 0.003 | 0.027 | 0.965 |
| 12 | 0.072 | 0.905 | 0.005 | 0.019 |
| 13 | 0.013 | 0.861 | 0.003 | 0.123 |
| 14 | 0.146 | 0.414 | 0.018 | 0.423 |
| 15 | 0.373 | 0.035 | 0.521 | 0.071 |
| 16 | 0.052 | 0.03 | 0.695 | 0.222 |
| 17 | 0.847 | 0.125 | 0.011 | 0.018 |
| 18 | 0.93 | 0.012 | 0.028 | 0.03 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.041 | 0.25 | -0.441 | 0.106 |
| 02 | -0.061 | 0.397 | -0.279 | -0.204 |
| 03 | 0.022 | 0.096 | -0.459 | 0.219 |
| 04 | -0.045 | 0.06 | -0.135 | 0.103 |
| 05 | -0.228 | 0.235 | -0.087 | 0.022 |
| 06 | -0.577 | 0.346 | -0.305 | 0.253 |
| 07 | -0.711 | 0.572 | 0.018 | -0.332 |
| 08 | 0.392 | 0.037 | -0.263 | -0.337 |
| 09 | -0.894 | 0.191 | -1.722 | 0.789 |
| 10 | -3.001 | -2.728 | -3.638 | 1.35 |
| 11 | -3.5 | -3.991 | -2.187 | 1.345 |
| 12 | -1.231 | 1.281 | -3.638 | -2.513 |
| 13 | -2.855 | 1.232 | -3.991 | -0.703 |
| 14 | -0.536 | 0.502 | -2.563 | 0.523 |
| 15 | 0.397 | -1.915 | 0.731 | -1.245 |
| 16 | -1.543 | -2.057 | 1.018 | -0.117 |
| 17 | 1.215 | -0.687 | -3.001 | -2.563 |
| 18 | 1.309 | -2.925 | -2.153 | -2.057 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.21246 |
| 0.0005 | 5.19821 |
| 0.0001 | 7.21821 |