| Motif | SOX8.H12CORE.0.PSM.A |
| Gene (human) | SOX8 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Sox8 |
| Gene synonyms (mouse) | Sox-8 |
| LOGO | 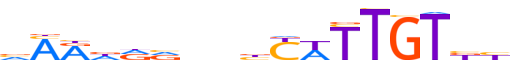 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | A |
| Motif | SOX8.H12CORE.0.PSM.A |
| Gene (human) | SOX8 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Sox8 |
| Gene synonyms (mouse) | Sox-8 |
| LOGO | 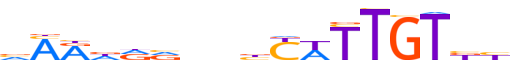 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 17 |
| Consensus | vMRddvnnbYWTTGTbb |
| GC content | 40.84% |
| Information content (bits; total / per base) | 11.418 / 0.672 |
| Data sources | ChIP-Seq + HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 1000 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 3 (15) | 0.789 | 0.838 | 0.726 | 0.774 | 0.782 | 0.831 | 2.85 | 3.105 | 116.208 | 165.0 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.988 | 0.983 | 0.967 | 0.957 | 0.914 | 0.907 |
| best | 1.0 | 0.999 | 0.999 | 0.999 | 0.998 | 0.998 | |
| Methyl HT-SELEX, 2 experiments | median | 0.989 | 0.983 | 0.968 | 0.958 | 0.916 | 0.91 |
| best | 1.0 | 0.999 | 0.999 | 0.999 | 0.998 | 0.998 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.973 | 0.963 | 0.932 | 0.919 | 0.868 | 0.865 |
| best | 1.0 | 0.999 | 0.998 | 0.998 | 0.994 | 0.992 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.791 | 0.528 | 0.676 | 0.455 |
| TF superclass | Other all-alpha-helical DNA-binding domains {4} (TFClass) |
| TF class | High-mobility group (HMG) domain factors {4.1} (TFClass) |
| TF family | SOX-related {4.1.1} (TFClass) |
| TF subfamily | Group E {4.1.1.5} (TFClass) |
| TFClass ID | TFClass: 4.1.1.5.1 |
| HGNC | HGNC:11203 |
| MGI | MGI:98370 |
| EntrezGene (human) | GeneID:30812 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:20681 (SSTAR profile) |
| UniProt ID (human) | SOX8_HUMAN |
| UniProt ID (mouse) | SOX8_MOUSE |
| UniProt AC (human) | P57073 (TFClass) |
| UniProt AC (mouse) | Q04886 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 3 human, 0 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 2 |
| PCM | SOX8.H12CORE.0.PSM.A.pcm |
| PWM | SOX8.H12CORE.0.PSM.A.pwm |
| PFM | SOX8.H12CORE.0.PSM.A.pfm |
| Alignment | SOX8.H12CORE.0.PSM.A.words.tsv |
| Threshold to P-value map | SOX8.H12CORE.0.PSM.A.thr |
| Motif in other formats | |
| JASPAR format | SOX8.H12CORE.0.PSM.A_jaspar_format.txt |
| MEME format | SOX8.H12CORE.0.PSM.A_meme_format.meme |
| Transfac format | SOX8.H12CORE.0.PSM.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 387.0 | 367.0 | 165.0 | 81.0 |
| 02 | 768.0 | 130.0 | 51.0 | 51.0 |
| 03 | 695.0 | 56.0 | 164.0 | 85.0 |
| 04 | 444.0 | 47.0 | 223.0 | 286.0 |
| 05 | 334.0 | 72.0 | 498.0 | 96.0 |
| 06 | 299.0 | 158.0 | 498.0 | 45.0 |
| 07 | 353.0 | 213.0 | 282.0 | 152.0 |
| 08 | 185.0 | 240.0 | 352.0 | 223.0 |
| 09 | 69.0 | 410.0 | 211.0 | 310.0 |
| 10 | 46.0 | 737.0 | 57.0 | 160.0 |
| 11 | 505.0 | 49.0 | 17.0 | 429.0 |
| 12 | 8.0 | 83.0 | 44.0 | 865.0 |
| 13 | 4.0 | 20.0 | 7.0 | 969.0 |
| 14 | 20.0 | 39.0 | 932.0 | 9.0 |
| 15 | 43.0 | 9.0 | 7.0 | 941.0 |
| 16 | 78.0 | 193.0 | 205.0 | 524.0 |
| 17 | 86.0 | 314.0 | 92.0 | 508.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.387 | 0.367 | 0.165 | 0.081 |
| 02 | 0.768 | 0.13 | 0.051 | 0.051 |
| 03 | 0.695 | 0.056 | 0.164 | 0.085 |
| 04 | 0.444 | 0.047 | 0.223 | 0.286 |
| 05 | 0.334 | 0.072 | 0.498 | 0.096 |
| 06 | 0.299 | 0.158 | 0.498 | 0.045 |
| 07 | 0.353 | 0.213 | 0.282 | 0.152 |
| 08 | 0.185 | 0.24 | 0.352 | 0.223 |
| 09 | 0.069 | 0.41 | 0.211 | 0.31 |
| 10 | 0.046 | 0.737 | 0.057 | 0.16 |
| 11 | 0.505 | 0.049 | 0.017 | 0.429 |
| 12 | 0.008 | 0.083 | 0.044 | 0.865 |
| 13 | 0.004 | 0.02 | 0.007 | 0.969 |
| 14 | 0.02 | 0.039 | 0.932 | 0.009 |
| 15 | 0.043 | 0.009 | 0.007 | 0.941 |
| 16 | 0.078 | 0.193 | 0.205 | 0.524 |
| 17 | 0.086 | 0.314 | 0.092 | 0.508 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.435 | 0.382 | -0.412 | -1.113 |
| 02 | 1.118 | -0.648 | -1.563 | -1.563 |
| 03 | 1.018 | -1.473 | -0.418 | -1.066 |
| 04 | 0.571 | -1.642 | -0.113 | 0.134 |
| 05 | 0.288 | -1.228 | 0.686 | -0.946 |
| 06 | 0.178 | -0.455 | 0.686 | -1.684 |
| 07 | 0.343 | -0.159 | 0.12 | -0.493 |
| 08 | -0.299 | -0.041 | 0.34 | -0.113 |
| 09 | -1.27 | 0.492 | -0.168 | 0.214 |
| 10 | -1.663 | 1.077 | -1.455 | -0.442 |
| 11 | 0.7 | -1.602 | -2.598 | 0.537 |
| 12 | -3.253 | -1.089 | -1.706 | 1.236 |
| 13 | -3.783 | -2.45 | -3.362 | 1.35 |
| 14 | -2.45 | -1.821 | 1.311 | -3.156 |
| 15 | -1.728 | -3.156 | -3.362 | 1.32 |
| 16 | -1.15 | -0.257 | -0.197 | 0.736 |
| 17 | -1.054 | 0.227 | -0.988 | 0.706 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.38441 |
| 0.0005 | 5.29701 |
| 0.0001 | 7.17951 |