| Motif | SOX17.H12INVITRO.2.S.C |
| Gene (human) | SOX17 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Sox17 |
| Gene synonyms (mouse) | Sox-17 |
| LOGO | 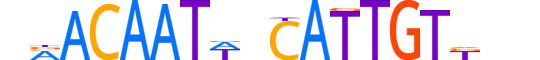 |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | C |
| Motif | SOX17.H12INVITRO.2.S.C |
| Gene (human) | SOX17 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Sox17 |
| Gene synonyms (mouse) | Sox-17 |
| LOGO | 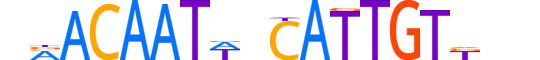 |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | C |
| Motif length | 18 |
| Consensus | nMACAATWnCATTGTKnn |
| GC content | 30.32% |
| Information content (bits; total / per base) | 22.079 / 1.227 |
| Data sources | HT-SELEX |
| Aligned words | 579 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 5 (31) | 0.539 | 0.553 | 0.372 | 0.393 | 0.538 | 0.558 | 1.197 | 1.291 | 4.398 | 6.854 |
| Mouse | 2 (10) | 0.513 | 0.524 | 0.361 | 0.371 | 0.471 | 0.499 | 1.075 | 1.151 | 9.83 | 12.155 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 2 experiments | median | 0.599 | 0.594 | 0.55 | 0.551 | 0.527 | 0.531 |
| best | 0.626 | 0.624 | 0.562 | 0.567 | 0.532 | 0.539 | |
| TF superclass | Other all-alpha-helical DNA-binding domains {4} (TFClass) |
| TF class | High-mobility group (HMG) domain factors {4.1} (TFClass) |
| TF family | SOX-related {4.1.1} (TFClass) |
| TF subfamily | Group F {4.1.1.6} (TFClass) |
| TFClass ID | TFClass: 4.1.1.6.2 |
| HGNC | HGNC:18122 |
| MGI | MGI:107543 |
| EntrezGene (human) | GeneID:64321 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:20671 (SSTAR profile) |
| UniProt ID (human) | SOX17_HUMAN |
| UniProt ID (mouse) | SOX17_MOUSE |
| UniProt AC (human) | Q9H6I2 (TFClass) |
| UniProt AC (mouse) | Q61473 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 5 human, 2 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 0 |
| PCM | SOX17.H12INVITRO.2.S.C.pcm |
| PWM | SOX17.H12INVITRO.2.S.C.pwm |
| PFM | SOX17.H12INVITRO.2.S.C.pfm |
| Alignment | SOX17.H12INVITRO.2.S.C.words.tsv |
| Threshold to P-value map | SOX17.H12INVITRO.2.S.C.thr |
| Motif in other formats | |
| JASPAR format | SOX17.H12INVITRO.2.S.C_jaspar_format.txt |
| MEME format | SOX17.H12INVITRO.2.S.C_meme_format.meme |
| Transfac format | SOX17.H12INVITRO.2.S.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 182.0 | 114.0 | 133.0 | 150.0 |
| 02 | 365.0 | 91.0 | 62.0 | 61.0 |
| 03 | 556.0 | 9.0 | 10.0 | 4.0 |
| 04 | 6.0 | 571.0 | 1.0 | 1.0 |
| 05 | 579.0 | 0.0 | 0.0 | 0.0 |
| 06 | 572.0 | 0.0 | 1.0 | 6.0 |
| 07 | 0.0 | 0.0 | 0.0 | 579.0 |
| 08 | 216.0 | 11.0 | 32.0 | 320.0 |
| 09 | 105.0 | 100.0 | 170.0 | 204.0 |
| 10 | 16.0 | 522.0 | 1.0 | 40.0 |
| 11 | 579.0 | 0.0 | 0.0 | 0.0 |
| 12 | 0.0 | 0.0 | 30.0 | 549.0 |
| 13 | 0.0 | 0.0 | 0.0 | 579.0 |
| 14 | 0.0 | 0.0 | 579.0 | 0.0 |
| 15 | 10.0 | 4.0 | 6.0 | 559.0 |
| 16 | 60.0 | 61.0 | 98.0 | 360.0 |
| 17 | 156.0 | 103.0 | 156.0 | 164.0 |
| 18 | 168.5 | 122.5 | 172.5 | 115.5 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.314 | 0.197 | 0.23 | 0.259 |
| 02 | 0.63 | 0.157 | 0.107 | 0.105 |
| 03 | 0.96 | 0.016 | 0.017 | 0.007 |
| 04 | 0.01 | 0.986 | 0.002 | 0.002 |
| 05 | 1.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.988 | 0.0 | 0.002 | 0.01 |
| 07 | 0.0 | 0.0 | 0.0 | 1.0 |
| 08 | 0.373 | 0.019 | 0.055 | 0.553 |
| 09 | 0.181 | 0.173 | 0.294 | 0.352 |
| 10 | 0.028 | 0.902 | 0.002 | 0.069 |
| 11 | 1.0 | 0.0 | 0.0 | 0.0 |
| 12 | 0.0 | 0.0 | 0.052 | 0.948 |
| 13 | 0.0 | 0.0 | 0.0 | 1.0 |
| 14 | 0.0 | 0.0 | 1.0 | 0.0 |
| 15 | 0.017 | 0.007 | 0.01 | 0.965 |
| 16 | 0.104 | 0.105 | 0.169 | 0.622 |
| 17 | 0.269 | 0.178 | 0.269 | 0.283 |
| 18 | 0.291 | 0.212 | 0.298 | 0.199 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.227 | -0.236 | -0.084 | 0.035 |
| 02 | 0.918 | -0.458 | -0.833 | -0.849 |
| 03 | 1.338 | -2.626 | -2.536 | -3.265 |
| 04 | -2.959 | 1.364 | -4.034 | -4.034 |
| 05 | 1.378 | -4.522 | -4.522 | -4.522 |
| 06 | 1.366 | -4.522 | -4.034 | -2.959 |
| 07 | -4.522 | -4.522 | -4.522 | 1.378 |
| 08 | 0.397 | -2.453 | -1.472 | 0.787 |
| 09 | -0.317 | -0.365 | 0.159 | 0.34 |
| 10 | -2.119 | 1.275 | -4.034 | -1.258 |
| 11 | 1.378 | -4.522 | -4.522 | -4.522 |
| 12 | -4.522 | -4.522 | -1.533 | 1.325 |
| 13 | -4.522 | -4.522 | -4.522 | 1.378 |
| 14 | -4.522 | -4.522 | 1.378 | -4.522 |
| 15 | -2.536 | -3.265 | -2.959 | 1.343 |
| 16 | -0.865 | -0.849 | -0.385 | 0.905 |
| 17 | 0.074 | -0.336 | 0.074 | 0.124 |
| 18 | 0.15 | -0.165 | 0.174 | -0.223 |
| P-value | Threshold |
|---|---|
| 0.001 | -2.53614 |
| 0.0005 | -0.80874 |
| 0.0001 | 2.86266 |