| Motif | SOX10.H12INVITRO.0.PSM.A |
| Gene (human) | SOX10 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Sox10 |
| Gene synonyms (mouse) | Sox-10, Sox21 |
| LOGO |  |
| LOGO (reverse complement) | 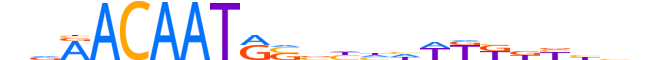 |
| Motif subtype | 0 |
| Quality | A |
| Motif | SOX10.H12INVITRO.0.PSM.A |
| Gene (human) | SOX10 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Sox10 |
| Gene synonyms (mouse) | Sox-10, Sox21 |
| LOGO |  |
| LOGO (reverse complement) | 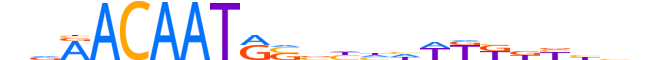 |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 22 |
| Consensus | nddRvMMWdddbSYATTGTYhn |
| GC content | 39.76% |
| Information content (bits; total / per base) | 16.2 / 0.736 |
| Data sources | ChIP-Seq + HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 9866 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (6) | 0.833 | 0.853 | 0.727 | 0.752 | 0.819 | 0.848 | 2.836 | 2.995 | 163.389 | 198.456 |
| Mouse | 1 (6) | 0.744 | 0.81 | 0.627 | 0.687 | 0.729 | 0.78 | 2.217 | 2.5 | 76.403 | 123.745 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 5 experiments | median | 0.996 | 0.995 | 0.902 | 0.908 | 0.723 | 0.757 |
| best | 0.999 | 0.999 | 0.998 | 0.997 | 0.932 | 0.938 | |
| Methyl HT-SELEX, 2 experiments | median | 0.999 | 0.999 | 0.95 | 0.952 | 0.828 | 0.847 |
| best | 0.999 | 0.999 | 0.998 | 0.997 | 0.932 | 0.938 | |
| Non-Methyl HT-SELEX, 3 experiments | median | 0.797 | 0.811 | 0.659 | 0.673 | 0.589 | 0.605 |
| best | 0.996 | 0.995 | 0.906 | 0.908 | 0.726 | 0.759 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.606 | 0.309 | 0.497 | 0.335 |
| TF superclass | Other all-alpha-helical DNA-binding domains {4} (TFClass) |
| TF class | High-mobility group (HMG) domain factors {4.1} (TFClass) |
| TF family | SOX-related {4.1.1} (TFClass) |
| TF subfamily | Group E {4.1.1.5} (TFClass) |
| TFClass ID | TFClass: 4.1.1.5.3 |
| HGNC | HGNC:11190 |
| MGI | MGI:98358 |
| EntrezGene (human) | GeneID:6663 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:20665 (SSTAR profile) |
| UniProt ID (human) | SOX10_HUMAN |
| UniProt ID (mouse) | SOX10_MOUSE |
| UniProt AC (human) | P56693 (TFClass) |
| UniProt AC (mouse) | Q04888 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 1 mouse |
| HT-SELEX | 3 |
| Methyl-HT-SELEX | 2 |
| PCM | SOX10.H12INVITRO.0.PSM.A.pcm |
| PWM | SOX10.H12INVITRO.0.PSM.A.pwm |
| PFM | SOX10.H12INVITRO.0.PSM.A.pfm |
| Alignment | SOX10.H12INVITRO.0.PSM.A.words.tsv |
| Threshold to P-value map | SOX10.H12INVITRO.0.PSM.A.thr |
| Motif in other formats | |
| JASPAR format | SOX10.H12INVITRO.0.PSM.A_jaspar_format.txt |
| MEME format | SOX10.H12INVITRO.0.PSM.A_meme_format.meme |
| Transfac format | SOX10.H12INVITRO.0.PSM.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 2573.5 | 2138.5 | 3355.5 | 1798.5 |
| 02 | 3178.0 | 1546.0 | 3504.0 | 1638.0 |
| 03 | 4441.0 | 1005.0 | 3038.0 | 1382.0 |
| 04 | 5993.0 | 997.0 | 1793.0 | 1083.0 |
| 05 | 4823.0 | 3302.0 | 919.0 | 822.0 |
| 06 | 5453.0 | 3399.0 | 621.0 | 393.0 |
| 07 | 6862.0 | 1998.0 | 448.0 | 558.0 |
| 08 | 6109.0 | 447.0 | 454.0 | 2856.0 |
| 09 | 3543.0 | 353.0 | 2434.0 | 3536.0 |
| 10 | 2439.0 | 508.0 | 4410.0 | 2509.0 |
| 11 | 2711.0 | 897.0 | 5071.0 | 1187.0 |
| 12 | 1417.0 | 1958.0 | 4725.0 | 1766.0 |
| 13 | 204.0 | 4897.0 | 3659.0 | 1106.0 |
| 14 | 78.0 | 5707.0 | 179.0 | 3902.0 |
| 15 | 9770.0 | 17.0 | 54.0 | 25.0 |
| 16 | 21.0 | 3.0 | 25.0 | 9817.0 |
| 17 | 2.0 | 23.0 | 16.0 | 9825.0 |
| 18 | 14.0 | 19.0 | 9832.0 | 1.0 |
| 19 | 59.0 | 14.0 | 20.0 | 9773.0 |
| 20 | 425.0 | 1102.0 | 875.0 | 7464.0 |
| 21 | 1593.75 | 4638.75 | 1309.75 | 2323.75 |
| 22 | 1888.25 | 2158.25 | 2420.25 | 3399.25 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.261 | 0.217 | 0.34 | 0.182 |
| 02 | 0.322 | 0.157 | 0.355 | 0.166 |
| 03 | 0.45 | 0.102 | 0.308 | 0.14 |
| 04 | 0.607 | 0.101 | 0.182 | 0.11 |
| 05 | 0.489 | 0.335 | 0.093 | 0.083 |
| 06 | 0.553 | 0.345 | 0.063 | 0.04 |
| 07 | 0.696 | 0.203 | 0.045 | 0.057 |
| 08 | 0.619 | 0.045 | 0.046 | 0.289 |
| 09 | 0.359 | 0.036 | 0.247 | 0.358 |
| 10 | 0.247 | 0.051 | 0.447 | 0.254 |
| 11 | 0.275 | 0.091 | 0.514 | 0.12 |
| 12 | 0.144 | 0.198 | 0.479 | 0.179 |
| 13 | 0.021 | 0.496 | 0.371 | 0.112 |
| 14 | 0.008 | 0.578 | 0.018 | 0.395 |
| 15 | 0.99 | 0.002 | 0.005 | 0.003 |
| 16 | 0.002 | 0.0 | 0.003 | 0.995 |
| 17 | 0.0 | 0.002 | 0.002 | 0.996 |
| 18 | 0.001 | 0.002 | 0.997 | 0.0 |
| 19 | 0.006 | 0.001 | 0.002 | 0.991 |
| 20 | 0.043 | 0.112 | 0.089 | 0.757 |
| 21 | 0.162 | 0.47 | 0.133 | 0.236 |
| 22 | 0.191 | 0.219 | 0.245 | 0.345 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.042 | -0.143 | 0.308 | -0.316 |
| 02 | 0.253 | -0.467 | 0.351 | -0.409 |
| 03 | 0.588 | -0.896 | 0.208 | -0.579 |
| 04 | 0.887 | -0.904 | -0.319 | -0.822 |
| 05 | 0.67 | 0.291 | -0.986 | -1.097 |
| 06 | 0.793 | 0.32 | -1.376 | -1.832 |
| 07 | 1.023 | -0.21 | -1.702 | -1.483 |
| 08 | 0.906 | -1.704 | -1.688 | 0.146 |
| 09 | 0.362 | -1.939 | -0.013 | 0.36 |
| 10 | -0.011 | -1.576 | 0.581 | 0.017 |
| 11 | 0.094 | -1.01 | 0.72 | -0.73 |
| 12 | -0.554 | -0.231 | 0.65 | -0.334 |
| 13 | -2.482 | 0.685 | 0.394 | -0.801 |
| 14 | -3.426 | 0.838 | -2.611 | 0.458 |
| 15 | 1.376 | -4.851 | -3.781 | -4.505 |
| 16 | -4.663 | -6.144 | -4.505 | 1.381 |
| 17 | -6.353 | -4.581 | -4.905 | 1.381 |
| 18 | -5.02 | -4.753 | 1.382 | -6.618 |
| 19 | -3.696 | -5.02 | -4.707 | 1.376 |
| 20 | -1.754 | -0.805 | -1.035 | 1.107 |
| 21 | -0.436 | 0.631 | -0.632 | -0.06 |
| 22 | -0.267 | -0.133 | -0.019 | 0.32 |
| P-value | Threshold |
|---|---|
| 0.001 | 1.42966 |
| 0.0005 | 2.90571 |
| 0.0001 | 6.00686 |