| Motif | SMAD3.H12CORE.2.P.C |
| Gene (human) | SMAD3 (GeneCards) |
| Gene synonyms (human) | MADH3 |
| Gene (mouse) | Smad3 |
| Gene synonyms (mouse) | Madh3 |
| LOGO | 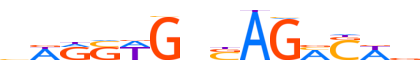 |
| LOGO (reverse complement) | 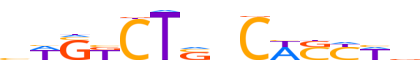 |
| Motif subtype | 2 |
| Quality | C |
| Motif | SMAD3.H12CORE.2.P.C |
| Gene (human) | SMAD3 (GeneCards) |
| Gene synonyms (human) | MADH3 |
| Gene (mouse) | Smad3 |
| Gene synonyms (mouse) | Madh3 |
| LOGO | 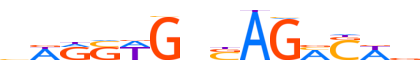 |
| LOGO (reverse complement) | 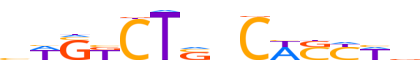 |
| Motif subtype | 2 |
| Quality | C |
| Motif length | 14 |
| Consensus | vWRSWGnSAGMSWv |
| GC content | 55.94% |
| Information content (bits; total / per base) | 9.914 / 0.708 |
| Data sources | ChIP-Seq |
| Aligned words | 1027 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 4 (24) | 0.614 | 0.742 | 0.417 | 0.6 | 0.629 | 0.744 | 1.509 | 2.357 | 8.488 | 55.509 |
| Mouse | 1 (5) | 0.582 | 0.585 | 0.404 | 0.411 | 0.602 | 0.613 | 1.465 | 1.488 | 1.886 | 3.658 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 2 experiments | median | 0.621 | 0.583 | 0.578 | 0.557 | 0.547 | 0.538 |
| best | 0.64 | 0.598 | 0.589 | 0.566 | 0.555 | 0.545 | |
| TF superclass | beta-Hairpin exposed by an alpha/beta-scaffold {7} (TFClass) |
| TF class | SMAD/NF-1 DNA-binding domain factors {7.1} (TFClass) |
| TF family | SMAD {7.1.1} (TFClass) |
| TF subfamily | R-MAD {7.1.1.1} (TFClass) |
| TFClass ID | TFClass: 7.1.1.1.3 |
| HGNC | HGNC:6769 |
| MGI | MGI:1201674 |
| EntrezGene (human) | GeneID:4088 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:17127 (SSTAR profile) |
| UniProt ID (human) | SMAD3_HUMAN |
| UniProt ID (mouse) | SMAD3_MOUSE |
| UniProt AC (human) | P84022 (TFClass) |
| UniProt AC (mouse) | Q8BUN5 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 4 human, 1 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 0 |
| PCM | SMAD3.H12CORE.2.P.C.pcm |
| PWM | SMAD3.H12CORE.2.P.C.pwm |
| PFM | SMAD3.H12CORE.2.P.C.pfm |
| Alignment | SMAD3.H12CORE.2.P.C.words.tsv |
| Threshold to P-value map | SMAD3.H12CORE.2.P.C.thr |
| Motif in other formats | |
| JASPAR format | SMAD3.H12CORE.2.P.C_jaspar_format.txt |
| MEME format | SMAD3.H12CORE.2.P.C_meme_format.meme |
| Transfac format | SMAD3.H12CORE.2.P.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 216.0 | 431.0 | 228.0 | 152.0 |
| 02 | 669.0 | 59.0 | 146.0 | 153.0 |
| 03 | 167.0 | 46.0 | 700.0 | 114.0 |
| 04 | 52.0 | 278.0 | 661.0 | 36.0 |
| 05 | 288.0 | 27.0 | 66.0 | 646.0 |
| 06 | 19.0 | 33.0 | 955.0 | 20.0 |
| 07 | 313.0 | 289.0 | 242.0 | 183.0 |
| 08 | 61.0 | 598.0 | 254.0 | 114.0 |
| 09 | 977.0 | 2.0 | 12.0 | 36.0 |
| 10 | 33.0 | 28.0 | 948.0 | 18.0 |
| 11 | 642.0 | 206.0 | 129.0 | 50.0 |
| 12 | 54.0 | 791.0 | 104.0 | 78.0 |
| 13 | 637.0 | 56.0 | 83.0 | 251.0 |
| 14 | 199.0 | 221.0 | 450.0 | 157.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.21 | 0.42 | 0.222 | 0.148 |
| 02 | 0.651 | 0.057 | 0.142 | 0.149 |
| 03 | 0.163 | 0.045 | 0.682 | 0.111 |
| 04 | 0.051 | 0.271 | 0.644 | 0.035 |
| 05 | 0.28 | 0.026 | 0.064 | 0.629 |
| 06 | 0.019 | 0.032 | 0.93 | 0.019 |
| 07 | 0.305 | 0.281 | 0.236 | 0.178 |
| 08 | 0.059 | 0.582 | 0.247 | 0.111 |
| 09 | 0.951 | 0.002 | 0.012 | 0.035 |
| 10 | 0.032 | 0.027 | 0.923 | 0.018 |
| 11 | 0.625 | 0.201 | 0.126 | 0.049 |
| 12 | 0.053 | 0.77 | 0.101 | 0.076 |
| 13 | 0.62 | 0.055 | 0.081 | 0.244 |
| 14 | 0.194 | 0.215 | 0.438 | 0.153 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.172 | 0.515 | -0.118 | -0.52 |
| 02 | 0.954 | -1.448 | -0.559 | -0.513 |
| 03 | -0.427 | -1.689 | 0.999 | -0.804 |
| 04 | -1.571 | 0.079 | 0.942 | -1.924 |
| 05 | 0.114 | -2.197 | -1.339 | 0.919 |
| 06 | -2.523 | -2.007 | 1.309 | -2.476 |
| 07 | 0.197 | 0.118 | -0.059 | -0.336 |
| 08 | -1.416 | 0.842 | -0.011 | -0.804 |
| 09 | 1.331 | -4.237 | -2.935 | -1.924 |
| 10 | -2.007 | -2.163 | 1.301 | -2.573 |
| 11 | 0.912 | -0.219 | -0.682 | -1.609 |
| 12 | -1.534 | 1.121 | -0.894 | -1.176 |
| 13 | 0.905 | -1.499 | -1.115 | -0.022 |
| 14 | -0.253 | -0.149 | 0.558 | -0.488 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.72251 |
| 0.0005 | 5.53091 |
| 0.0001 | 7.19446 |