| Motif | SMAD2.H12INVITRO.0.P.D |
| Gene (human) | SMAD2 (GeneCards) |
| Gene synonyms (human) | MADH2, MADR2 |
| Gene (mouse) | Smad2 |
| Gene synonyms (mouse) | Madh2, Madr2 |
| LOGO |  |
| LOGO (reverse complement) | 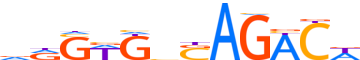 |
| Motif subtype | 0 |
| Quality | D |
| Motif | SMAD2.H12INVITRO.0.P.D |
| Gene (human) | SMAD2 (GeneCards) |
| Gene synonyms (human) | MADH2, MADR2 |
| Gene (mouse) | Smad2 |
| Gene synonyms (mouse) | Madh2, Madr2 |
| LOGO |  |
| LOGO (reverse complement) | 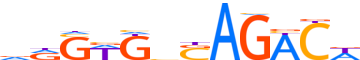 |
| Motif subtype | 0 |
| Quality | D |
| Motif length | 12 |
| Consensus | WGTCTSdMWSYh |
| GC content | 53.81% |
| Information content (bits; total / per base) | 10.148 / 0.846 |
| Data sources | ChIP-Seq |
| Aligned words | 367 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 10 (67) | 0.788 | 0.841 | 0.705 | 0.783 | 0.782 | 0.839 | 3.047 | 3.566 | 68.77 | 118.409 |
| Mouse | 6 (41) | 0.655 | 0.736 | 0.491 | 0.618 | 0.654 | 0.731 | 1.78 | 2.429 | 14.398 | 74.638 |
| TF superclass | beta-Hairpin exposed by an alpha/beta-scaffold {7} (TFClass) |
| TF class | SMAD/NF-1 DNA-binding domain factors {7.1} (TFClass) |
| TF family | SMAD {7.1.1} (TFClass) |
| TF subfamily | R-MAD {7.1.1.1} (TFClass) |
| TFClass ID | TFClass: 7.1.1.1.2 |
| HGNC | HGNC:6768 |
| MGI | MGI:108051 |
| EntrezGene (human) | GeneID:4087 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:17126 (SSTAR profile) |
| UniProt ID (human) | SMAD2_HUMAN |
| UniProt ID (mouse) | SMAD2_MOUSE |
| UniProt AC (human) | Q15796 (TFClass) |
| UniProt AC (mouse) | Q62432 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 10 human, 6 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | SMAD2.H12INVITRO.0.P.D.pcm |
| PWM | SMAD2.H12INVITRO.0.P.D.pwm |
| PFM | SMAD2.H12INVITRO.0.P.D.pfm |
| Alignment | SMAD2.H12INVITRO.0.P.D.words.tsv |
| Threshold to P-value map | SMAD2.H12INVITRO.0.P.D.thr |
| Motif in other formats | |
| JASPAR format | SMAD2.H12INVITRO.0.P.D_jaspar_format.txt |
| MEME format | SMAD2.H12INVITRO.0.P.D_meme_format.meme |
| Transfac format | SMAD2.H12INVITRO.0.P.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 131.0 | 37.0 | 16.0 | 183.0 |
| 02 | 16.0 | 14.0 | 329.0 | 8.0 |
| 03 | 25.0 | 20.0 | 23.0 | 299.0 |
| 04 | 1.0 | 345.0 | 10.0 | 11.0 |
| 05 | 1.0 | 2.0 | 1.0 | 363.0 |
| 06 | 50.0 | 69.0 | 238.0 | 10.0 |
| 07 | 91.0 | 47.0 | 144.0 | 85.0 |
| 08 | 29.0 | 295.0 | 21.0 | 22.0 |
| 09 | 244.0 | 33.0 | 24.0 | 66.0 |
| 10 | 17.0 | 291.0 | 37.0 | 22.0 |
| 11 | 49.0 | 212.0 | 23.0 | 83.0 |
| 12 | 60.0 | 94.0 | 45.0 | 168.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.357 | 0.101 | 0.044 | 0.499 |
| 02 | 0.044 | 0.038 | 0.896 | 0.022 |
| 03 | 0.068 | 0.054 | 0.063 | 0.815 |
| 04 | 0.003 | 0.94 | 0.027 | 0.03 |
| 05 | 0.003 | 0.005 | 0.003 | 0.989 |
| 06 | 0.136 | 0.188 | 0.649 | 0.027 |
| 07 | 0.248 | 0.128 | 0.392 | 0.232 |
| 08 | 0.079 | 0.804 | 0.057 | 0.06 |
| 09 | 0.665 | 0.09 | 0.065 | 0.18 |
| 10 | 0.046 | 0.793 | 0.101 | 0.06 |
| 11 | 0.134 | 0.578 | 0.063 | 0.226 |
| 12 | 0.163 | 0.256 | 0.123 | 0.458 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.351 | -0.885 | -1.674 | 0.682 |
| 02 | -1.674 | -1.796 | 1.266 | -2.286 |
| 03 | -1.259 | -1.468 | -1.337 | 1.17 |
| 04 | -3.628 | 1.313 | -2.095 | -2.011 |
| 05 | -3.628 | -3.289 | -3.628 | 1.363 |
| 06 | -0.594 | -0.28 | 0.943 | -2.095 |
| 07 | -0.008 | -0.654 | 0.445 | -0.075 |
| 08 | -1.118 | 1.157 | -1.423 | -1.379 |
| 09 | 0.968 | -0.995 | -1.297 | -0.323 |
| 10 | -1.619 | 1.143 | -0.885 | -1.379 |
| 11 | -0.614 | 0.828 | -1.337 | -0.099 |
| 12 | -0.416 | 0.024 | -0.696 | 0.598 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.72616 |
| 0.0005 | 5.53617 |
| 0.0001 | 7.20937 |