| Motif | SIX2.H12RSNP.0.P.B |
| Gene (human) | SIX2 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Six2 |
| Gene synonyms (mouse) | |
| LOGO | 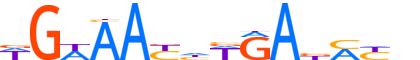 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | SIX2.H12RSNP.0.P.B |
| Gene (human) | SIX2 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Six2 |
| Gene synonyms (mouse) | |
| LOGO | 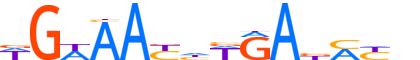 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 14 |
| Consensus | WGWAAYhWRAbMYb |
| GC content | 38.89% |
| Information content (bits; total / per base) | 12.089 / 0.864 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 5 (34) | 0.904 | 0.929 | 0.814 | 0.864 | 0.914 | 0.936 | 3.53 | 3.961 | 248.168 | 300.569 |
| Mouse | 8 (52) | 0.892 | 0.928 | 0.812 | 0.866 | 0.892 | 0.929 | 3.364 | 3.829 | 211.571 | 334.174 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.686 | 0.659 | 0.629 | 0.61 | 0.58 | 0.574 |
| best | 0.872 | 0.834 | 0.77 | 0.74 | 0.667 | 0.661 | |
| Methyl HT-SELEX, 2 experiments | median | 0.704 | 0.679 | 0.648 | 0.628 | 0.592 | 0.586 |
| best | 0.872 | 0.834 | 0.77 | 0.74 | 0.667 | 0.661 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.683 | 0.656 | 0.627 | 0.609 | 0.578 | 0.572 |
| best | 0.836 | 0.793 | 0.733 | 0.704 | 0.643 | 0.635 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 27.273 | 3.862 | 0.443 | 0.36 |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.767 | 0.509 | 0.812 | 0.635 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | HD-SINE {3.1.6} (TFClass) |
| TF subfamily | SIX1-like {3.1.6.1} (TFClass) |
| TFClass ID | TFClass: 3.1.6.1.2 |
| HGNC | HGNC:10888 |
| MGI | MGI:102778 |
| EntrezGene (human) | GeneID:10736 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:20472 (SSTAR profile) |
| UniProt ID (human) | SIX2_HUMAN |
| UniProt ID (mouse) | SIX2_MOUSE |
| UniProt AC (human) | Q9NPC8 (TFClass) |
| UniProt AC (mouse) | Q62232 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 5 human, 8 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 2 |
| PCM | SIX2.H12RSNP.0.P.B.pcm |
| PWM | SIX2.H12RSNP.0.P.B.pwm |
| PFM | SIX2.H12RSNP.0.P.B.pfm |
| Alignment | SIX2.H12RSNP.0.P.B.words.tsv |
| Threshold to P-value map | SIX2.H12RSNP.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | SIX2.H12RSNP.0.P.B_jaspar_format.txt |
| MEME format | SIX2.H12RSNP.0.P.B_meme_format.meme |
| Transfac format | SIX2.H12RSNP.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 215.0 | 11.0 | 196.0 | 578.0 |
| 02 | 12.0 | 4.0 | 982.0 | 2.0 |
| 03 | 515.0 | 37.0 | 42.0 | 406.0 |
| 04 | 898.0 | 59.0 | 9.0 | 34.0 |
| 05 | 992.0 | 2.0 | 5.0 | 1.0 |
| 06 | 41.0 | 532.0 | 64.0 | 363.0 |
| 07 | 238.0 | 409.0 | 63.0 | 290.0 |
| 08 | 130.0 | 90.0 | 97.0 | 683.0 |
| 09 | 177.0 | 15.0 | 766.0 | 42.0 |
| 10 | 991.0 | 2.0 | 3.0 | 4.0 |
| 11 | 79.0 | 174.0 | 251.0 | 496.0 |
| 12 | 517.0 | 425.0 | 21.0 | 37.0 |
| 13 | 87.0 | 558.0 | 48.0 | 307.0 |
| 14 | 139.0 | 367.0 | 212.0 | 282.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.215 | 0.011 | 0.196 | 0.578 |
| 02 | 0.012 | 0.004 | 0.982 | 0.002 |
| 03 | 0.515 | 0.037 | 0.042 | 0.406 |
| 04 | 0.898 | 0.059 | 0.009 | 0.034 |
| 05 | 0.992 | 0.002 | 0.005 | 0.001 |
| 06 | 0.041 | 0.532 | 0.064 | 0.363 |
| 07 | 0.238 | 0.409 | 0.063 | 0.29 |
| 08 | 0.13 | 0.09 | 0.097 | 0.683 |
| 09 | 0.177 | 0.015 | 0.766 | 0.042 |
| 10 | 0.991 | 0.002 | 0.003 | 0.004 |
| 11 | 0.079 | 0.174 | 0.251 | 0.496 |
| 12 | 0.517 | 0.425 | 0.021 | 0.037 |
| 13 | 0.087 | 0.558 | 0.048 | 0.307 |
| 14 | 0.139 | 0.367 | 0.212 | 0.282 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.15 | -2.985 | -0.241 | 0.834 |
| 02 | -2.909 | -3.783 | 1.363 | -4.213 |
| 03 | 0.719 | -1.872 | -1.75 | 0.482 |
| 04 | 1.274 | -1.422 | -3.156 | -1.952 |
| 05 | 1.373 | -4.213 | -3.622 | -4.525 |
| 06 | -1.774 | 0.752 | -1.343 | 0.371 |
| 07 | -0.049 | 0.49 | -1.358 | 0.147 |
| 08 | -0.648 | -1.01 | -0.936 | 1.001 |
| 09 | -0.342 | -2.711 | 1.115 | -1.75 |
| 10 | 1.372 | -4.213 | -3.975 | -3.783 |
| 11 | -1.137 | -0.359 | 0.004 | 0.682 |
| 12 | 0.723 | 0.528 | -2.405 | -1.872 |
| 13 | -1.043 | 0.799 | -1.622 | 0.204 |
| 14 | -0.582 | 0.382 | -0.164 | 0.12 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.09281 |
| 0.0005 | 5.14861 |
| 0.0001 | 7.26411 |