| Motif | SIX2.H12INVIVO.2.SM.B |
| Gene (human) | SIX2 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Six2 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 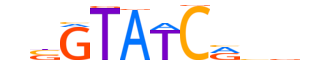 |
| Motif subtype | 2 |
| Quality | B |
| Motif | SIX2.H12INVIVO.2.SM.B |
| Gene (human) | SIX2 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Six2 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | B |
| Motif length | 11 |
| Consensus | nhvYGATACvn |
| GC content | 49.87% |
| Information content (bits; total / per base) | 9.037 / 0.822 |
| Data sources | HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 2431 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 5 (34) | 0.634 | 0.664 | 0.455 | 0.495 | 0.586 | 0.615 | 1.323 | 1.418 | 24.577 | 35.62 |
| Mouse | 8 (52) | 0.639 | 0.669 | 0.46 | 0.504 | 0.586 | 0.616 | 1.321 | 1.465 | 37.234 | 51.398 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.887 | 0.832 | 0.834 | 0.78 | 0.766 | 0.722 |
| best | 0.914 | 0.869 | 0.86 | 0.813 | 0.78 | 0.745 | |
| Methyl HT-SELEX, 2 experiments | median | 0.901 | 0.851 | 0.851 | 0.799 | 0.777 | 0.737 |
| best | 0.914 | 0.869 | 0.86 | 0.813 | 0.78 | 0.745 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.88 | 0.825 | 0.826 | 0.772 | 0.754 | 0.714 |
| best | 0.887 | 0.831 | 0.827 | 0.775 | 0.76 | 0.715 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 1.561 | 0.262 | 0.266 | 0.029 |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.667 | 0.299 | 0.494 | 0.358 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | HD-SINE {3.1.6} (TFClass) |
| TF subfamily | SIX1-like {3.1.6.1} (TFClass) |
| TFClass ID | TFClass: 3.1.6.1.2 |
| HGNC | HGNC:10888 |
| MGI | MGI:102778 |
| EntrezGene (human) | GeneID:10736 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:20472 (SSTAR profile) |
| UniProt ID (human) | SIX2_HUMAN |
| UniProt ID (mouse) | SIX2_MOUSE |
| UniProt AC (human) | Q9NPC8 (TFClass) |
| UniProt AC (mouse) | Q62232 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 5 human, 8 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 2 |
| PCM | SIX2.H12INVIVO.2.SM.B.pcm |
| PWM | SIX2.H12INVIVO.2.SM.B.pwm |
| PFM | SIX2.H12INVIVO.2.SM.B.pfm |
| Alignment | SIX2.H12INVIVO.2.SM.B.words.tsv |
| Threshold to P-value map | SIX2.H12INVIVO.2.SM.B.thr |
| Motif in other formats | |
| JASPAR format | SIX2.H12INVIVO.2.SM.B_jaspar_format.txt |
| MEME format | SIX2.H12INVIVO.2.SM.B_meme_format.meme |
| Transfac format | SIX2.H12INVIVO.2.SM.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 782.25 | 486.25 | 478.25 | 684.25 |
| 02 | 594.25 | 1008.25 | 411.25 | 417.25 |
| 03 | 693.0 | 424.0 | 931.0 | 383.0 |
| 04 | 46.0 | 1138.0 | 265.0 | 982.0 |
| 05 | 3.0 | 56.0 | 2372.0 | 0.0 |
| 06 | 1944.0 | 8.0 | 1.0 | 478.0 |
| 07 | 27.0 | 69.0 | 0.0 | 2335.0 |
| 08 | 2348.0 | 83.0 | 0.0 | 0.0 |
| 09 | 25.0 | 2106.0 | 136.0 | 164.0 |
| 10 | 210.75 | 1062.75 | 1009.75 | 147.75 |
| 11 | 403.5 | 802.5 | 488.5 | 736.5 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.322 | 0.2 | 0.197 | 0.281 |
| 02 | 0.244 | 0.415 | 0.169 | 0.172 |
| 03 | 0.285 | 0.174 | 0.383 | 0.158 |
| 04 | 0.019 | 0.468 | 0.109 | 0.404 |
| 05 | 0.001 | 0.023 | 0.976 | 0.0 |
| 06 | 0.8 | 0.003 | 0.0 | 0.197 |
| 07 | 0.011 | 0.028 | 0.0 | 0.961 |
| 08 | 0.966 | 0.034 | 0.0 | 0.0 |
| 09 | 0.01 | 0.866 | 0.056 | 0.067 |
| 10 | 0.087 | 0.437 | 0.415 | 0.061 |
| 11 | 0.166 | 0.33 | 0.201 | 0.303 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.252 | -0.222 | -0.239 | 0.118 |
| 02 | -0.022 | 0.505 | -0.389 | -0.375 |
| 03 | 0.131 | -0.359 | 0.425 | -0.46 |
| 04 | -2.543 | 0.626 | -0.826 | 0.479 |
| 05 | -4.814 | -2.353 | 1.359 | -5.746 |
| 06 | 1.161 | -4.115 | -5.331 | -0.239 |
| 07 | -3.047 | -2.151 | -5.746 | 1.344 |
| 08 | 1.349 | -1.971 | -5.746 | -5.746 |
| 09 | -3.119 | 1.241 | -1.486 | -1.301 |
| 10 | -1.053 | 0.557 | 0.506 | -1.404 |
| 11 | -0.408 | 0.277 | -0.218 | 0.192 |
| P-value | Threshold |
|---|---|
| 0.001 | 5.08725 |
| 0.0005 | 5.970165 |
| 0.0001 | 7.50814 |