| Motif | SIX2.H12INVIVO.0.P.B |
| Gene (human) | SIX2 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Six2 |
| Gene synonyms (mouse) | |
| LOGO | 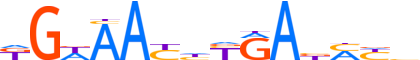 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | SIX2.H12INVIVO.0.P.B |
| Gene (human) | SIX2 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Six2 |
| Gene synonyms (mouse) | |
| LOGO | 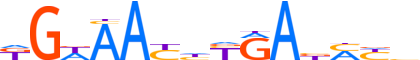 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 14 |
| Consensus | KGWAAYhYRAbMYb |
| GC content | 40.82% |
| Information content (bits; total / per base) | 11.933 / 0.852 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 5 (34) | 0.913 | 0.935 | 0.829 | 0.872 | 0.914 | 0.935 | 3.494 | 3.904 | 244.071 | 287.796 |
| Mouse | 8 (52) | 0.901 | 0.936 | 0.828 | 0.882 | 0.898 | 0.934 | 3.437 | 3.902 | 220.284 | 340.745 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.679 | 0.654 | 0.625 | 0.607 | 0.578 | 0.572 |
| best | 0.867 | 0.829 | 0.764 | 0.734 | 0.664 | 0.657 | |
| Methyl HT-SELEX, 2 experiments | median | 0.695 | 0.673 | 0.641 | 0.623 | 0.589 | 0.583 |
| best | 0.867 | 0.829 | 0.764 | 0.734 | 0.664 | 0.657 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.676 | 0.651 | 0.623 | 0.606 | 0.576 | 0.571 |
| best | 0.834 | 0.791 | 0.732 | 0.702 | 0.643 | 0.635 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 27.273 | 3.862 | 0.433 | 0.313 |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.744 | 0.504 | 0.798 | 0.612 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | HD-SINE {3.1.6} (TFClass) |
| TF subfamily | SIX1-like {3.1.6.1} (TFClass) |
| TFClass ID | TFClass: 3.1.6.1.2 |
| HGNC | HGNC:10888 |
| MGI | MGI:102778 |
| EntrezGene (human) | GeneID:10736 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:20472 (SSTAR profile) |
| UniProt ID (human) | SIX2_HUMAN |
| UniProt ID (mouse) | SIX2_MOUSE |
| UniProt AC (human) | Q9NPC8 (TFClass) |
| UniProt AC (mouse) | Q62232 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 5 human, 8 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 2 |
| PCM | SIX2.H12INVIVO.0.P.B.pcm |
| PWM | SIX2.H12INVIVO.0.P.B.pwm |
| PFM | SIX2.H12INVIVO.0.P.B.pfm |
| Alignment | SIX2.H12INVIVO.0.P.B.words.tsv |
| Threshold to P-value map | SIX2.H12INVIVO.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | SIX2.H12INVIVO.0.P.B_jaspar_format.txt |
| MEME format | SIX2.H12INVIVO.0.P.B_meme_format.meme |
| Transfac format | SIX2.H12INVIVO.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 196.0 | 7.0 | 261.0 | 536.0 |
| 02 | 7.0 | 1.0 | 981.0 | 11.0 |
| 03 | 479.0 | 43.0 | 70.0 | 408.0 |
| 04 | 888.0 | 61.0 | 4.0 | 47.0 |
| 05 | 997.0 | 1.0 | 2.0 | 0.0 |
| 06 | 38.0 | 546.0 | 57.0 | 359.0 |
| 07 | 162.0 | 415.0 | 63.0 | 360.0 |
| 08 | 99.0 | 118.0 | 104.0 | 679.0 |
| 09 | 166.0 | 12.0 | 764.0 | 58.0 |
| 10 | 996.0 | 1.0 | 3.0 | 0.0 |
| 11 | 63.0 | 180.0 | 309.0 | 448.0 |
| 12 | 501.0 | 411.0 | 23.0 | 65.0 |
| 13 | 101.0 | 587.0 | 75.0 | 237.0 |
| 14 | 148.0 | 423.0 | 193.0 | 236.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.196 | 0.007 | 0.261 | 0.536 |
| 02 | 0.007 | 0.001 | 0.981 | 0.011 |
| 03 | 0.479 | 0.043 | 0.07 | 0.408 |
| 04 | 0.888 | 0.061 | 0.004 | 0.047 |
| 05 | 0.997 | 0.001 | 0.002 | 0.0 |
| 06 | 0.038 | 0.546 | 0.057 | 0.359 |
| 07 | 0.162 | 0.415 | 0.063 | 0.36 |
| 08 | 0.099 | 0.118 | 0.104 | 0.679 |
| 09 | 0.166 | 0.012 | 0.764 | 0.058 |
| 10 | 0.996 | 0.001 | 0.003 | 0.0 |
| 11 | 0.063 | 0.18 | 0.309 | 0.448 |
| 12 | 0.501 | 0.411 | 0.023 | 0.065 |
| 13 | 0.101 | 0.587 | 0.075 | 0.237 |
| 14 | 0.148 | 0.423 | 0.193 | 0.236 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.241 | -3.362 | 0.043 | 0.759 |
| 02 | -3.362 | -4.525 | 1.362 | -2.985 |
| 03 | 0.647 | -1.728 | -1.255 | 0.487 |
| 04 | 1.263 | -1.39 | -3.783 | -1.642 |
| 05 | 1.378 | -4.525 | -4.213 | -4.982 |
| 06 | -1.846 | 0.777 | -1.455 | 0.36 |
| 07 | -0.43 | 0.504 | -1.358 | 0.363 |
| 08 | -0.916 | -0.743 | -0.867 | 0.995 |
| 09 | -0.406 | -2.909 | 1.112 | -1.439 |
| 10 | 1.377 | -4.525 | -3.975 | -4.982 |
| 11 | -1.358 | -0.326 | 0.211 | 0.58 |
| 12 | 0.692 | 0.494 | -2.32 | -1.328 |
| 13 | -0.896 | 0.85 | -1.188 | -0.053 |
| 14 | -0.52 | 0.523 | -0.257 | -0.057 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.17661 |
| 0.0005 | 5.23411 |
| 0.0001 | 7.30851 |