| Motif | SIX2.H12CORE.0.P.B |
| Gene (human) | SIX2 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Six2 |
| Gene synonyms (mouse) | |
| LOGO | 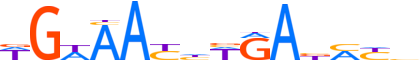 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | SIX2.H12CORE.0.P.B |
| Gene (human) | SIX2 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Six2 |
| Gene synonyms (mouse) | |
| LOGO | 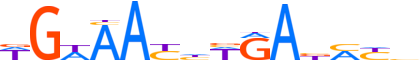 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 14 |
| Consensus | WGWAAYhWRAbMYb |
| GC content | 39.16% |
| Information content (bits; total / per base) | 11.799 / 0.843 |
| Data sources | ChIP-Seq |
| Aligned words | 1003 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 5 (34) | 0.909 | 0.933 | 0.82 | 0.869 | 0.916 | 0.937 | 3.55 | 3.997 | 246.914 | 294.638 |
| Mouse | 8 (52) | 0.898 | 0.933 | 0.821 | 0.874 | 0.896 | 0.933 | 3.399 | 3.887 | 216.237 | 349.097 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.689 | 0.662 | 0.633 | 0.613 | 0.583 | 0.576 |
| best | 0.873 | 0.836 | 0.77 | 0.74 | 0.668 | 0.661 | |
| Methyl HT-SELEX, 2 experiments | median | 0.706 | 0.681 | 0.649 | 0.629 | 0.594 | 0.587 |
| best | 0.873 | 0.836 | 0.77 | 0.74 | 0.668 | 0.661 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.686 | 0.659 | 0.631 | 0.611 | 0.581 | 0.575 |
| best | 0.84 | 0.798 | 0.737 | 0.708 | 0.647 | 0.639 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 27.273 | 3.862 | 0.441 | 0.341 |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.761 | 0.514 | 0.808 | 0.624 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | HD-SINE {3.1.6} (TFClass) |
| TF subfamily | SIX1-like {3.1.6.1} (TFClass) |
| TFClass ID | TFClass: 3.1.6.1.2 |
| HGNC | HGNC:10888 |
| MGI | MGI:102778 |
| EntrezGene (human) | GeneID:10736 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:20472 (SSTAR profile) |
| UniProt ID (human) | SIX2_HUMAN |
| UniProt ID (mouse) | SIX2_MOUSE |
| UniProt AC (human) | Q9NPC8 (TFClass) |
| UniProt AC (mouse) | Q62232 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 5 human, 8 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 2 |
| PCM | SIX2.H12CORE.0.P.B.pcm |
| PWM | SIX2.H12CORE.0.P.B.pwm |
| PFM | SIX2.H12CORE.0.P.B.pfm |
| Alignment | SIX2.H12CORE.0.P.B.words.tsv |
| Threshold to P-value map | SIX2.H12CORE.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | SIX2.H12CORE.0.P.B_jaspar_format.txt |
| MEME format | SIX2.H12CORE.0.P.B_meme_format.meme |
| Transfac format | SIX2.H12CORE.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 206.0 | 10.0 | 193.0 | 594.0 |
| 02 | 15.0 | 4.0 | 982.0 | 2.0 |
| 03 | 471.0 | 42.0 | 55.0 | 435.0 |
| 04 | 883.0 | 65.0 | 10.0 | 45.0 |
| 05 | 999.0 | 0.0 | 1.0 | 3.0 |
| 06 | 41.0 | 520.0 | 49.0 | 393.0 |
| 07 | 211.0 | 396.0 | 61.0 | 335.0 |
| 08 | 121.0 | 120.0 | 98.0 | 664.0 |
| 09 | 166.0 | 20.0 | 766.0 | 51.0 |
| 10 | 989.0 | 10.0 | 2.0 | 2.0 |
| 11 | 80.0 | 194.0 | 241.0 | 488.0 |
| 12 | 516.0 | 403.0 | 34.0 | 50.0 |
| 13 | 94.0 | 560.0 | 55.0 | 294.0 |
| 14 | 126.0 | 402.0 | 206.0 | 269.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.205 | 0.01 | 0.192 | 0.592 |
| 02 | 0.015 | 0.004 | 0.979 | 0.002 |
| 03 | 0.47 | 0.042 | 0.055 | 0.434 |
| 04 | 0.88 | 0.065 | 0.01 | 0.045 |
| 05 | 0.996 | 0.0 | 0.001 | 0.003 |
| 06 | 0.041 | 0.518 | 0.049 | 0.392 |
| 07 | 0.21 | 0.395 | 0.061 | 0.334 |
| 08 | 0.121 | 0.12 | 0.098 | 0.662 |
| 09 | 0.166 | 0.02 | 0.764 | 0.051 |
| 10 | 0.986 | 0.01 | 0.002 | 0.002 |
| 11 | 0.08 | 0.193 | 0.24 | 0.487 |
| 12 | 0.514 | 0.402 | 0.034 | 0.05 |
| 13 | 0.094 | 0.558 | 0.055 | 0.293 |
| 14 | 0.126 | 0.401 | 0.205 | 0.268 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.195 | -3.069 | -0.26 | 0.858 |
| 02 | -2.714 | -3.786 | 1.36 | -4.216 |
| 03 | 0.627 | -1.753 | -1.493 | 0.548 |
| 04 | 1.254 | -1.331 | -3.069 | -1.687 |
| 05 | 1.377 | -4.985 | -4.528 | -3.978 |
| 06 | -1.776 | 0.726 | -1.605 | 0.447 |
| 07 | -0.171 | 0.454 | -1.393 | 0.288 |
| 08 | -0.721 | -0.73 | -0.929 | 0.97 |
| 09 | -0.409 | -2.453 | 1.112 | -1.566 |
| 10 | 1.367 | -3.069 | -4.216 | -4.216 |
| 11 | -1.128 | -0.255 | -0.039 | 0.663 |
| 12 | 0.718 | 0.472 | -1.955 | -1.585 |
| 13 | -0.97 | 0.8 | -1.493 | 0.158 |
| 14 | -0.681 | 0.469 | -0.195 | 0.07 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.23061 |
| 0.0005 | 5.25566 |
| 0.0001 | 7.29082 |