| Motif | SIX1.H12CORE.0.P.B |
| Gene (human) | SIX1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Six1 |
| Gene synonyms (mouse) | |
| LOGO | 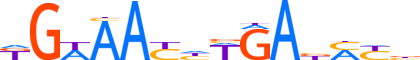 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | SIX1.H12CORE.0.P.B |
| Gene (human) | SIX1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Six1 |
| Gene synonyms (mouse) | |
| LOGO | 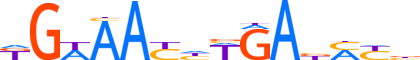 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 14 |
| Consensus | KGWAAYhWGAbMYb |
| GC content | 40.2% |
| Information content (bits; total / per base) | 12.84 / 0.917 |
| Data sources | ChIP-Seq |
| Aligned words | 1001 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 3 (18) | 0.927 | 0.954 | 0.88 | 0.917 | 0.929 | 0.956 | 4.108 | 4.534 | 270.535 | 343.538 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.691 | 0.669 | 0.625 | 0.611 | 0.575 | 0.572 |
| best | 0.868 | 0.843 | 0.724 | 0.717 | 0.625 | 0.634 | |
| Methyl HT-SELEX, 2 experiments | median | 0.699 | 0.682 | 0.621 | 0.615 | 0.566 | 0.571 |
| best | 0.868 | 0.843 | 0.724 | 0.717 | 0.622 | 0.634 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.691 | 0.669 | 0.625 | 0.611 | 0.576 | 0.572 |
| best | 0.835 | 0.811 | 0.711 | 0.698 | 0.625 | 0.627 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.752 | 0.439 | 0.766 | 0.64 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | HD-SINE {3.1.6} (TFClass) |
| TF subfamily | SIX1-like {3.1.6.1} (TFClass) |
| TFClass ID | TFClass: 3.1.6.1.1 |
| HGNC | HGNC:10887 |
| MGI | MGI:102780 |
| EntrezGene (human) | GeneID:6495 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:20471 (SSTAR profile) |
| UniProt ID (human) | SIX1_HUMAN |
| UniProt ID (mouse) | SIX1_MOUSE |
| UniProt AC (human) | Q15475 (TFClass) |
| UniProt AC (mouse) | Q62231 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 3 human, 0 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 2 |
| PCM | SIX1.H12CORE.0.P.B.pcm |
| PWM | SIX1.H12CORE.0.P.B.pwm |
| PFM | SIX1.H12CORE.0.P.B.pfm |
| Alignment | SIX1.H12CORE.0.P.B.words.tsv |
| Threshold to P-value map | SIX1.H12CORE.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | SIX1.H12CORE.0.P.B_jaspar_format.txt |
| MEME format | SIX1.H12CORE.0.P.B_meme_format.meme |
| Transfac format | SIX1.H12CORE.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 196.0 | 20.0 | 204.0 | 581.0 |
| 02 | 5.0 | 2.0 | 992.0 | 2.0 |
| 03 | 508.0 | 36.0 | 34.0 | 423.0 |
| 04 | 906.0 | 60.0 | 4.0 | 31.0 |
| 05 | 999.0 | 1.0 | 1.0 | 0.0 |
| 06 | 35.0 | 556.0 | 43.0 | 367.0 |
| 07 | 182.0 | 449.0 | 54.0 | 316.0 |
| 08 | 104.0 | 95.0 | 88.0 | 714.0 |
| 09 | 104.0 | 7.0 | 845.0 | 45.0 |
| 10 | 1000.0 | 0.0 | 1.0 | 0.0 |
| 11 | 77.0 | 155.0 | 271.0 | 498.0 |
| 12 | 519.0 | 405.0 | 22.0 | 55.0 |
| 13 | 70.0 | 578.0 | 59.0 | 294.0 |
| 14 | 104.0 | 417.0 | 234.0 | 246.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.196 | 0.02 | 0.204 | 0.58 |
| 02 | 0.005 | 0.002 | 0.991 | 0.002 |
| 03 | 0.507 | 0.036 | 0.034 | 0.423 |
| 04 | 0.905 | 0.06 | 0.004 | 0.031 |
| 05 | 0.998 | 0.001 | 0.001 | 0.0 |
| 06 | 0.035 | 0.555 | 0.043 | 0.367 |
| 07 | 0.182 | 0.449 | 0.054 | 0.316 |
| 08 | 0.104 | 0.095 | 0.088 | 0.713 |
| 09 | 0.104 | 0.007 | 0.844 | 0.045 |
| 10 | 0.999 | 0.0 | 0.001 | 0.0 |
| 11 | 0.077 | 0.155 | 0.271 | 0.498 |
| 12 | 0.518 | 0.405 | 0.022 | 0.055 |
| 13 | 0.07 | 0.577 | 0.059 | 0.294 |
| 14 | 0.104 | 0.417 | 0.234 | 0.246 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.242 | -2.451 | -0.203 | 0.838 |
| 02 | -3.623 | -4.214 | 1.372 | -4.214 |
| 03 | 0.705 | -1.899 | -1.953 | 0.522 |
| 04 | 1.282 | -1.407 | -3.784 | -2.041 |
| 05 | 1.379 | -4.526 | -4.526 | -4.983 |
| 06 | -1.926 | 0.795 | -1.729 | 0.381 |
| 07 | -0.316 | 0.582 | -1.509 | 0.232 |
| 08 | -0.868 | -0.957 | -1.033 | 1.044 |
| 09 | -0.868 | -3.363 | 1.212 | -1.685 |
| 10 | 1.38 | -4.983 | -4.526 | -4.983 |
| 11 | -1.163 | -0.475 | 0.079 | 0.685 |
| 12 | 0.726 | 0.479 | -2.363 | -1.491 |
| 13 | -1.256 | 0.833 | -1.423 | 0.16 |
| 14 | -0.868 | 0.508 | -0.067 | -0.017 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.76896 |
| 0.0005 | 4.90651 |
| 0.0001 | 7.16626 |