| Motif | RUNX2.H12INVIVO.2.M.C |
| Gene (human) | RUNX2 (GeneCards) |
| Gene synonyms (human) | AML3, CBFA1, OSF2, PEBP2A |
| Gene (mouse) | Runx2 |
| Gene synonyms (mouse) | Aml3, Cbfa1, Osf2, Pebp2a |
| LOGO | 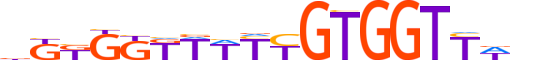 |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | C |
| Motif | RUNX2.H12INVIVO.2.M.C |
| Gene (human) | RUNX2 (GeneCards) |
| Gene synonyms (human) | AML3, CBFA1, OSF2, PEBP2A |
| Gene (mouse) | Runx2 |
| Gene synonyms (mouse) | Aml3, Cbfa1, Osf2, Pebp2a |
| LOGO |  |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | C |
| Motif length | 18 |
| Consensus | bKKGKKWWYYGTGGTYWn |
| GC content | 45.93% |
| Information content (bits; total / per base) | 18.561 / 1.031 |
| Data sources | Methyl-HT-SELEX |
| Aligned words | 5894 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 4 (26) | 0.858 | 0.899 | 0.734 | 0.801 | 0.836 | 0.894 | 2.773 | 3.572 | 76.753 | 160.886 |
| Mouse | 16 (101) | 0.809 | 0.878 | 0.666 | 0.774 | 0.806 | 0.867 | 2.643 | 3.057 | 78.602 | 134.638 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 3 experiments | median | 0.77 | 0.727 | 0.667 | 0.646 | 0.594 | 0.593 |
| best | 0.993 | 0.991 | 0.952 | 0.943 | 0.821 | 0.827 | |
| Methyl HT-SELEX, 1 experiments | median | 0.993 | 0.991 | 0.952 | 0.943 | 0.821 | 0.827 |
| best | 0.993 | 0.991 | 0.952 | 0.943 | 0.821 | 0.827 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.728 | 0.688 | 0.639 | 0.62 | 0.579 | 0.576 |
| best | 0.77 | 0.727 | 0.667 | 0.646 | 0.594 | 0.593 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 2.475 | 1.827 | 0.192 | 0.128 |
| TF superclass | Immunoglobulin fold {6} (TFClass) |
| TF class | Runt domain factors {6.4} (TFClass) |
| TF family | Runt-related {6.4.1} (TFClass) |
| TF subfamily | {6.4.1.0} (TFClass) |
| TFClass ID | TFClass: 6.4.1.0.1 |
| HGNC | HGNC:10472 |
| MGI | MGI:99829 |
| EntrezGene (human) | GeneID:860 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:12393 (SSTAR profile) |
| UniProt ID (human) | RUNX2_HUMAN |
| UniProt ID (mouse) | RUNX2_MOUSE |
| UniProt AC (human) | Q13950 (TFClass) |
| UniProt AC (mouse) | Q08775 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 4 human, 16 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 1 |
| PCM | RUNX2.H12INVIVO.2.M.C.pcm |
| PWM | RUNX2.H12INVIVO.2.M.C.pwm |
| PFM | RUNX2.H12INVIVO.2.M.C.pfm |
| Alignment | RUNX2.H12INVIVO.2.M.C.words.tsv |
| Threshold to P-value map | RUNX2.H12INVIVO.2.M.C.thr |
| Motif in other formats | |
| JASPAR format | RUNX2.H12INVIVO.2.M.C_jaspar_format.txt |
| MEME format | RUNX2.H12INVIVO.2.M.C_meme_format.meme |
| Transfac format | RUNX2.H12INVIVO.2.M.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 565.0 | 1602.0 | 1662.0 | 2065.0 |
| 02 | 156.75 | 365.75 | 4131.75 | 1239.75 |
| 03 | 197.0 | 503.0 | 1663.0 | 3531.0 |
| 04 | 143.0 | 260.0 | 4792.0 | 699.0 |
| 05 | 222.0 | 244.0 | 4306.0 | 1122.0 |
| 06 | 377.0 | 306.0 | 907.0 | 4304.0 |
| 07 | 525.0 | 292.0 | 497.0 | 4580.0 |
| 08 | 1137.0 | 339.0 | 245.0 | 4173.0 |
| 09 | 392.0 | 699.0 | 241.0 | 4562.0 |
| 10 | 55.0 | 2574.0 | 48.0 | 3217.0 |
| 11 | 8.0 | 2.0 | 5881.0 | 3.0 |
| 12 | 26.0 | 270.0 | 3.0 | 5595.0 |
| 13 | 1.0 | 0.0 | 5893.0 | 0.0 |
| 14 | 0.0 | 0.0 | 5893.0 | 1.0 |
| 15 | 3.0 | 50.0 | 63.0 | 5778.0 |
| 16 | 278.0 | 879.0 | 146.0 | 4591.0 |
| 17 | 2521.75 | 170.75 | 559.75 | 2641.75 |
| 18 | 1161.0 | 1456.0 | 1781.0 | 1496.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.096 | 0.272 | 0.282 | 0.35 |
| 02 | 0.027 | 0.062 | 0.701 | 0.21 |
| 03 | 0.033 | 0.085 | 0.282 | 0.599 |
| 04 | 0.024 | 0.044 | 0.813 | 0.119 |
| 05 | 0.038 | 0.041 | 0.731 | 0.19 |
| 06 | 0.064 | 0.052 | 0.154 | 0.73 |
| 07 | 0.089 | 0.05 | 0.084 | 0.777 |
| 08 | 0.193 | 0.058 | 0.042 | 0.708 |
| 09 | 0.067 | 0.119 | 0.041 | 0.774 |
| 10 | 0.009 | 0.437 | 0.008 | 0.546 |
| 11 | 0.001 | 0.0 | 0.998 | 0.001 |
| 12 | 0.004 | 0.046 | 0.001 | 0.949 |
| 13 | 0.0 | 0.0 | 1.0 | 0.0 |
| 14 | 0.0 | 0.0 | 1.0 | 0.0 |
| 15 | 0.001 | 0.008 | 0.011 | 0.98 |
| 16 | 0.047 | 0.149 | 0.025 | 0.779 |
| 17 | 0.428 | 0.029 | 0.095 | 0.448 |
| 18 | 0.197 | 0.247 | 0.302 | 0.254 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.956 | 0.083 | 0.12 | 0.337 |
| 02 | -2.228 | -1.389 | 1.03 | -0.172 |
| 03 | -2.003 | -1.072 | 0.121 | 0.873 |
| 04 | -2.319 | -1.728 | 1.178 | -0.744 |
| 05 | -1.884 | -1.791 | 1.071 | -0.272 |
| 06 | -1.359 | -1.566 | -0.484 | 1.071 |
| 07 | -1.029 | -1.613 | -1.084 | 1.133 |
| 08 | -0.259 | -1.464 | -1.787 | 1.04 |
| 09 | -1.32 | -0.744 | -1.803 | 1.129 |
| 10 | -3.251 | 0.557 | -3.381 | 0.78 |
| 11 | -4.977 | -5.869 | 1.383 | -5.654 |
| 12 | -3.959 | -1.69 | -5.654 | 1.333 |
| 13 | -6.143 | -6.522 | 1.385 | -6.522 |
| 14 | -6.522 | -6.522 | 1.385 | -6.143 |
| 15 | -5.654 | -3.342 | -3.12 | 1.365 |
| 16 | -1.661 | -0.516 | -2.299 | 1.135 |
| 17 | 0.537 | -2.144 | -0.966 | 0.583 |
| 18 | -0.238 | -0.012 | 0.189 | 0.015 |
| P-value | Threshold |
|---|---|
| 0.001 | 0.20626 |
| 0.0005 | 1.80381 |
| 0.0001 | 5.08556 |