| Motif | RARG.H12RSNP.3.S.B |
| Gene (human) | RARG (GeneCards) |
| Gene synonyms (human) | NR1B3 |
| Gene (mouse) | Rarg |
| Gene synonyms (mouse) | Nr1b3 |
| LOGO |  |
| LOGO (reverse complement) | 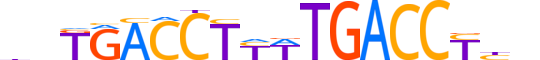 |
| Motif subtype | 3 |
| Quality | B |
| Motif | RARG.H12RSNP.3.S.B |
| Gene (human) | RARG (GeneCards) |
| Gene synonyms (human) | NR1B3 |
| Gene (mouse) | Rarg |
| Gene synonyms (mouse) | Nr1b3 |
| LOGO |  |
| LOGO (reverse complement) |  |
| Motif subtype | 3 |
| Quality | B |
| Motif length | 18 |
| Consensus | nvRGGTCAWRAGGTCAnd |
| GC content | 47.42% |
| Information content (bits; total / per base) | 20.853 / 1.159 |
| Data sources | HT-SELEX |
| Aligned words | 1019 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 5 (21) | 0.596 | 0.641 | 0.412 | 0.444 | 0.554 | 0.606 | 1.35 | 1.685 | 10.42 | 27.131 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 10 experiments | median | 0.867 | 0.856 | 0.714 | 0.717 | 0.613 | 0.631 |
| best | 1.0 | 1.0 | 0.998 | 0.997 | 0.986 | 0.983 | |
| Methyl HT-SELEX, 2 experiments | median | 0.976 | 0.963 | 0.959 | 0.941 | 0.924 | 0.904 |
| best | 0.999 | 0.999 | 0.996 | 0.995 | 0.984 | 0.98 | |
| Non-Methyl HT-SELEX, 8 experiments | median | 0.801 | 0.797 | 0.66 | 0.67 | 0.581 | 0.601 |
| best | 1.0 | 1.0 | 0.998 | 0.997 | 0.986 | 0.983 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.955 | 0.074 | 0.934 | 0.382 |
| batch 2 | 0.615 | 0.114 | 0.488 | 0.341 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Nuclear receptors with C4 zinc fingers {2.1} (TFClass) |
| TF family | Thyroid hormone receptor-related {2.1.2} (TFClass) |
| TF subfamily | RAR (NR1B) {2.1.2.1} (TFClass) |
| TFClass ID | TFClass: 2.1.2.1.3 |
| HGNC | HGNC:9866 |
| MGI | MGI:97858 |
| EntrezGene (human) | GeneID:5916 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:19411 (SSTAR profile) |
| UniProt ID (human) | RARG_HUMAN |
| UniProt ID (mouse) | RARG_MOUSE |
| UniProt AC (human) | P13631 (TFClass) |
| UniProt AC (mouse) | P18911 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 5 human, 0 mouse |
| HT-SELEX | 8 |
| Methyl-HT-SELEX | 2 |
| PCM | RARG.H12RSNP.3.S.B.pcm |
| PWM | RARG.H12RSNP.3.S.B.pwm |
| PFM | RARG.H12RSNP.3.S.B.pfm |
| Alignment | RARG.H12RSNP.3.S.B.words.tsv |
| Threshold to P-value map | RARG.H12RSNP.3.S.B.thr |
| Motif in other formats | |
| JASPAR format | RARG.H12RSNP.3.S.B_jaspar_format.txt |
| MEME format | RARG.H12RSNP.3.S.B_meme_format.meme |
| Transfac format | RARG.H12RSNP.3.S.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 273.5 | 300.5 | 173.5 | 271.5 |
| 02 | 313.25 | 100.25 | 506.25 | 99.25 |
| 03 | 746.0 | 55.0 | 209.0 | 9.0 |
| 04 | 0.0 | 0.0 | 1019.0 | 0.0 |
| 05 | 0.0 | 0.0 | 1006.0 | 13.0 |
| 06 | 0.0 | 0.0 | 0.0 | 1019.0 |
| 07 | 0.0 | 1015.0 | 1.0 | 3.0 |
| 08 | 1010.0 | 0.0 | 8.0 | 1.0 |
| 09 | 679.0 | 63.0 | 18.0 | 259.0 |
| 10 | 685.0 | 9.0 | 187.0 | 138.0 |
| 11 | 889.0 | 13.0 | 114.0 | 3.0 |
| 12 | 56.0 | 3.0 | 960.0 | 0.0 |
| 13 | 15.0 | 5.0 | 915.0 | 84.0 |
| 14 | 1.0 | 1.0 | 130.0 | 887.0 |
| 15 | 3.0 | 857.0 | 50.0 | 109.0 |
| 16 | 841.0 | 99.0 | 44.0 | 35.0 |
| 17 | 377.0 | 252.0 | 167.0 | 223.0 |
| 18 | 411.75 | 183.75 | 233.75 | 189.75 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.268 | 0.295 | 0.17 | 0.266 |
| 02 | 0.307 | 0.098 | 0.497 | 0.097 |
| 03 | 0.732 | 0.054 | 0.205 | 0.009 |
| 04 | 0.0 | 0.0 | 1.0 | 0.0 |
| 05 | 0.0 | 0.0 | 0.987 | 0.013 |
| 06 | 0.0 | 0.0 | 0.0 | 1.0 |
| 07 | 0.0 | 0.996 | 0.001 | 0.003 |
| 08 | 0.991 | 0.0 | 0.008 | 0.001 |
| 09 | 0.666 | 0.062 | 0.018 | 0.254 |
| 10 | 0.672 | 0.009 | 0.184 | 0.135 |
| 11 | 0.872 | 0.013 | 0.112 | 0.003 |
| 12 | 0.055 | 0.003 | 0.942 | 0.0 |
| 13 | 0.015 | 0.005 | 0.898 | 0.082 |
| 14 | 0.001 | 0.001 | 0.128 | 0.87 |
| 15 | 0.003 | 0.841 | 0.049 | 0.107 |
| 16 | 0.825 | 0.097 | 0.043 | 0.034 |
| 17 | 0.37 | 0.247 | 0.164 | 0.219 |
| 18 | 0.404 | 0.18 | 0.229 | 0.186 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.071 | 0.164 | -0.381 | 0.063 |
| 02 | 0.205 | -0.922 | 0.683 | -0.932 |
| 03 | 1.07 | -1.509 | -0.196 | -3.174 |
| 04 | -4.998 | -4.998 | 1.381 | -4.998 |
| 05 | -4.998 | -4.998 | 1.368 | -2.857 |
| 06 | -4.998 | -4.998 | -4.998 | 1.381 |
| 07 | -4.998 | 1.377 | -4.542 | -3.993 |
| 08 | 1.372 | -4.998 | -3.272 | -4.542 |
| 09 | 0.976 | -1.377 | -2.565 | 0.016 |
| 10 | 0.985 | -3.174 | -0.307 | -0.607 |
| 11 | 1.245 | -2.857 | -0.796 | -3.993 |
| 12 | -1.491 | -3.993 | 1.322 | -4.998 |
| 13 | -2.73 | -3.64 | 1.274 | -1.096 |
| 14 | -4.542 | -4.542 | -0.666 | 1.243 |
| 15 | -3.993 | 1.208 | -1.601 | -0.84 |
| 16 | 1.19 | -0.935 | -1.724 | -1.943 |
| 17 | 0.39 | -0.011 | -0.419 | -0.132 |
| 18 | 0.478 | -0.324 | -0.085 | -0.292 |
| P-value | Threshold |
|---|---|
| 0.001 | -1.48939 |
| 0.0005 | 0.18811 |
| 0.0001 | 3.72181 |