| Motif | RARB.H12INVIVO.1.P.B |
| Gene (human) | RARB (GeneCards) |
| Gene synonyms (human) | HAP, NR1B2 |
| Gene (mouse) | Rarb |
| Gene synonyms (mouse) | Nr1b2 |
| LOGO | 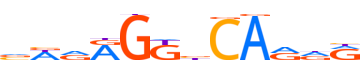 |
| LOGO (reverse complement) | 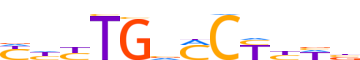 |
| Motif subtype | 1 |
| Quality | B |
| Motif | RARB.H12INVIVO.1.P.B |
| Gene (human) | RARB (GeneCards) |
| Gene synonyms (human) | HAP, NR1B2 |
| Gene (mouse) | Rarb |
| Gene synonyms (mouse) | Nr1b2 |
| LOGO | 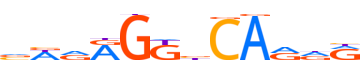 |
| LOGO (reverse complement) | 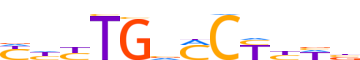 |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 12 |
| Consensus | vWdRGKbCARdR |
| GC content | 55.36% |
| Information content (bits; total / per base) | 8.624 / 0.719 |
| Data sources | ChIP-Seq |
| Aligned words | 1028 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | 2 (14) | 0.621 | 0.664 | 0.45 | 0.506 | 0.608 | 0.646 | 1.47 | 1.681 | 15.353 | 32.721 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.743 | 0.695 | 0.65 | 0.629 | 0.578 | 0.58 |
| best | 0.808 | 0.752 | 0.706 | 0.674 | 0.615 | 0.612 | |
| Methyl HT-SELEX, 1 experiments | median | 0.808 | 0.752 | 0.706 | 0.674 | 0.615 | 0.612 |
| best | 0.808 | 0.752 | 0.706 | 0.674 | 0.615 | 0.612 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.678 | 0.639 | 0.593 | 0.583 | 0.541 | 0.548 |
| best | 0.678 | 0.639 | 0.593 | 0.583 | 0.541 | 0.548 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Nuclear receptors with C4 zinc fingers {2.1} (TFClass) |
| TF family | Thyroid hormone receptor-related {2.1.2} (TFClass) |
| TF subfamily | RAR (NR1B) {2.1.2.1} (TFClass) |
| TFClass ID | TFClass: 2.1.2.1.2 |
| HGNC | HGNC:9865 |
| MGI | MGI:97857 |
| EntrezGene (human) | GeneID:5915 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:218772 (SSTAR profile) |
| UniProt ID (human) | RARB_HUMAN |
| UniProt ID (mouse) | RARB_MOUSE |
| UniProt AC (human) | P10826 (TFClass) |
| UniProt AC (mouse) | P22605 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 2 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| PCM | RARB.H12INVIVO.1.P.B.pcm |
| PWM | RARB.H12INVIVO.1.P.B.pwm |
| PFM | RARB.H12INVIVO.1.P.B.pfm |
| Alignment | RARB.H12INVIVO.1.P.B.words.tsv |
| Threshold to P-value map | RARB.H12INVIVO.1.P.B.thr |
| Motif in other formats | |
| JASPAR format | RARB.H12INVIVO.1.P.B_jaspar_format.txt |
| MEME format | RARB.H12INVIVO.1.P.B_meme_format.meme |
| Transfac format | RARB.H12INVIVO.1.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 179.0 | 475.0 | 267.0 | 107.0 |
| 02 | 628.0 | 119.0 | 135.0 | 146.0 |
| 03 | 367.0 | 89.0 | 475.0 | 97.0 |
| 04 | 683.0 | 7.0 | 242.0 | 96.0 |
| 05 | 18.0 | 12.0 | 969.0 | 29.0 |
| 06 | 50.0 | 45.0 | 698.0 | 235.0 |
| 07 | 132.0 | 217.0 | 198.0 | 481.0 |
| 08 | 18.0 | 946.0 | 49.0 | 15.0 |
| 09 | 960.0 | 12.0 | 45.0 | 11.0 |
| 10 | 407.0 | 68.0 | 494.0 | 59.0 |
| 11 | 405.0 | 76.0 | 445.0 | 102.0 |
| 12 | 191.0 | 73.0 | 673.0 | 91.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.174 | 0.462 | 0.26 | 0.104 |
| 02 | 0.611 | 0.116 | 0.131 | 0.142 |
| 03 | 0.357 | 0.087 | 0.462 | 0.094 |
| 04 | 0.664 | 0.007 | 0.235 | 0.093 |
| 05 | 0.018 | 0.012 | 0.943 | 0.028 |
| 06 | 0.049 | 0.044 | 0.679 | 0.229 |
| 07 | 0.128 | 0.211 | 0.193 | 0.468 |
| 08 | 0.018 | 0.92 | 0.048 | 0.015 |
| 09 | 0.934 | 0.012 | 0.044 | 0.011 |
| 10 | 0.396 | 0.066 | 0.481 | 0.057 |
| 11 | 0.394 | 0.074 | 0.433 | 0.099 |
| 12 | 0.186 | 0.071 | 0.655 | 0.089 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.359 | 0.611 | 0.038 | -0.867 |
| 02 | 0.889 | -0.762 | -0.638 | -0.56 |
| 03 | 0.354 | -1.048 | 0.611 | -0.963 |
| 04 | 0.973 | -3.389 | -0.06 | -0.974 |
| 05 | -2.573 | -2.936 | 1.322 | -2.13 |
| 06 | -1.61 | -1.711 | 0.995 | -0.089 |
| 07 | -0.66 | -0.168 | -0.259 | 0.624 |
| 08 | -2.573 | 1.298 | -1.629 | -2.738 |
| 09 | 1.313 | -2.936 | -1.711 | -3.012 |
| 10 | 0.457 | -1.311 | 0.65 | -1.449 |
| 11 | 0.452 | -1.203 | 0.546 | -0.914 |
| 12 | -0.294 | -1.242 | 0.959 | -1.026 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.966215 |
| 0.0005 | 5.701215 |
| 0.0001 | 7.17091 |