| Motif | PITX3.H12RSNP.1.S.B |
| Gene (human) | PITX3 (GeneCards) |
| Gene synonyms (human) | PTX3 |
| Gene (mouse) | Pitx3 |
| Gene synonyms (mouse) | |
| LOGO | 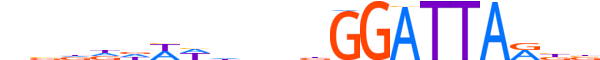 |
| LOGO (reverse complement) | 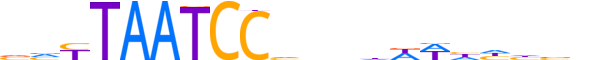 |
| Motif subtype | 1 |
| Quality | B |
| Motif | PITX3.H12RSNP.1.S.B |
| Gene (human) | PITX3 (GeneCards) |
| Gene synonyms (human) | PTX3 |
| Gene (mouse) | Pitx3 |
| Gene synonyms (mouse) | |
| LOGO | 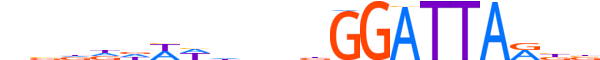 |
| LOGO (reverse complement) | 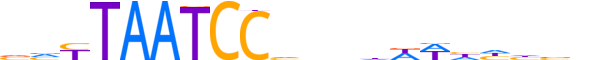 |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 20 |
| Consensus | nbdddWWhhnbGGATTARbb |
| GC content | 42.88% |
| Information content (bits; total / per base) | 14.911 / 0.746 |
| Data sources | HT-SELEX |
| Aligned words | 933 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 6 experiments | median | 0.975 | 0.957 | 0.954 | 0.927 | 0.871 | 0.859 |
| best | 0.994 | 0.991 | 0.975 | 0.968 | 0.94 | 0.915 | |
| Methyl HT-SELEX, 2 experiments | median | 0.97 | 0.947 | 0.956 | 0.929 | 0.919 | 0.89 |
| best | 0.977 | 0.959 | 0.968 | 0.946 | 0.94 | 0.915 | |
| Non-Methyl HT-SELEX, 4 experiments | median | 0.975 | 0.959 | 0.932 | 0.913 | 0.828 | 0.818 |
| best | 0.994 | 0.991 | 0.975 | 0.968 | 0.93 | 0.904 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.663 | 0.26 | 0.611 | 0.412 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | Paired-related HD {3.1.3} (TFClass) |
| TF subfamily | PITX {3.1.3.19} (TFClass) |
| TFClass ID | TFClass: 3.1.3.19.3 |
| HGNC | HGNC:9006 |
| MGI | MGI:1100498 |
| EntrezGene (human) | GeneID:5309 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:18742 (SSTAR profile) |
| UniProt ID (human) | PITX3_HUMAN |
| UniProt ID (mouse) | PITX3_MOUSE |
| UniProt AC (human) | O75364 (TFClass) |
| UniProt AC (mouse) | O35160 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 4 |
| Methyl-HT-SELEX | 2 |
| PCM | PITX3.H12RSNP.1.S.B.pcm |
| PWM | PITX3.H12RSNP.1.S.B.pwm |
| PFM | PITX3.H12RSNP.1.S.B.pfm |
| Alignment | PITX3.H12RSNP.1.S.B.words.tsv |
| Threshold to P-value map | PITX3.H12RSNP.1.S.B.thr |
| Motif in other formats | |
| JASPAR format | PITX3.H12RSNP.1.S.B_jaspar_format.txt |
| MEME format | PITX3.H12RSNP.1.S.B_meme_format.meme |
| Transfac format | PITX3.H12RSNP.1.S.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 162.75 | 269.75 | 332.75 | 167.75 |
| 02 | 143.75 | 211.75 | 428.75 | 148.75 |
| 03 | 194.0 | 110.0 | 443.0 | 186.0 |
| 04 | 258.0 | 22.0 | 398.0 | 255.0 |
| 05 | 330.0 | 24.0 | 234.0 | 345.0 |
| 06 | 460.0 | 54.0 | 27.0 | 392.0 |
| 07 | 314.0 | 94.0 | 47.0 | 478.0 |
| 08 | 304.0 | 202.0 | 97.0 | 330.0 |
| 09 | 333.0 | 287.0 | 156.0 | 157.0 |
| 10 | 279.0 | 270.0 | 255.0 | 129.0 |
| 11 | 98.0 | 211.0 | 365.0 | 259.0 |
| 12 | 19.0 | 7.0 | 895.0 | 12.0 |
| 13 | 0.0 | 2.0 | 930.0 | 1.0 |
| 14 | 909.0 | 23.0 | 1.0 | 0.0 |
| 15 | 0.0 | 2.0 | 0.0 | 931.0 |
| 16 | 0.0 | 1.0 | 1.0 | 931.0 |
| 17 | 926.0 | 1.0 | 1.0 | 5.0 |
| 18 | 512.0 | 41.0 | 343.0 | 37.0 |
| 19 | 67.0 | 144.0 | 407.0 | 315.0 |
| 20 | 116.25 | 195.25 | 468.25 | 153.25 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.174 | 0.289 | 0.357 | 0.18 |
| 02 | 0.154 | 0.227 | 0.46 | 0.159 |
| 03 | 0.208 | 0.118 | 0.475 | 0.199 |
| 04 | 0.277 | 0.024 | 0.427 | 0.273 |
| 05 | 0.354 | 0.026 | 0.251 | 0.37 |
| 06 | 0.493 | 0.058 | 0.029 | 0.42 |
| 07 | 0.337 | 0.101 | 0.05 | 0.512 |
| 08 | 0.326 | 0.217 | 0.104 | 0.354 |
| 09 | 0.357 | 0.308 | 0.167 | 0.168 |
| 10 | 0.299 | 0.289 | 0.273 | 0.138 |
| 11 | 0.105 | 0.226 | 0.391 | 0.278 |
| 12 | 0.02 | 0.008 | 0.959 | 0.013 |
| 13 | 0.0 | 0.002 | 0.997 | 0.001 |
| 14 | 0.974 | 0.025 | 0.001 | 0.0 |
| 15 | 0.0 | 0.002 | 0.0 | 0.998 |
| 16 | 0.0 | 0.001 | 0.001 | 0.998 |
| 17 | 0.992 | 0.001 | 0.001 | 0.005 |
| 18 | 0.549 | 0.044 | 0.368 | 0.04 |
| 19 | 0.072 | 0.154 | 0.436 | 0.338 |
| 20 | 0.125 | 0.209 | 0.502 | 0.164 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.357 | 0.144 | 0.353 | -0.327 |
| 02 | -0.48 | -0.096 | 0.605 | -0.446 |
| 03 | -0.183 | -0.744 | 0.638 | -0.225 |
| 04 | 0.1 | -2.294 | 0.531 | 0.089 |
| 05 | 0.345 | -2.213 | 0.003 | 0.389 |
| 06 | 0.676 | -1.439 | -2.102 | 0.516 |
| 07 | 0.295 | -0.898 | -1.574 | 0.714 |
| 08 | 0.263 | -0.143 | -0.867 | 0.345 |
| 09 | 0.354 | 0.206 | -0.399 | -0.392 |
| 10 | 0.178 | 0.145 | 0.089 | -0.586 |
| 11 | -0.857 | -0.099 | 0.445 | 0.104 |
| 12 | -2.429 | -3.295 | 1.339 | -2.841 |
| 13 | -4.923 | -4.148 | 1.378 | -4.463 |
| 14 | 1.355 | -2.252 | -4.463 | -4.923 |
| 15 | -4.923 | -4.148 | -4.923 | 1.379 |
| 16 | -4.923 | -4.463 | -4.463 | 1.379 |
| 17 | 1.373 | -4.463 | -4.463 | -3.556 |
| 18 | 0.782 | -1.705 | 0.383 | -1.803 |
| 19 | -1.23 | -0.478 | 0.554 | 0.299 |
| 20 | -0.689 | -0.176 | 0.693 | -0.416 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.35806 |
| 0.0005 | 3.71181 |
| 0.0001 | 6.49606 |