| Motif | PITX1.H12INVIVO.1.S.B |
| Gene (human) | PITX1 (GeneCards) |
| Gene synonyms (human) | BFT, PTX1 |
| Gene (mouse) | Pitx1 |
| Gene synonyms (mouse) | Bft, Potx, Ptx1 |
| LOGO | 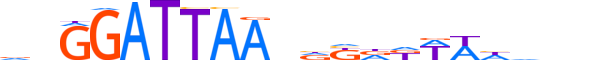 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | B |
| Motif | PITX1.H12INVIVO.1.S.B |
| Gene (human) | PITX1 (GeneCards) |
| Gene synonyms (human) | BFT, PTX1 |
| Gene (mouse) | Pitx1 |
| Gene synonyms (mouse) | Bft, Potx, Ptx1 |
| LOGO | 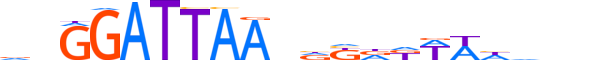 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 20 |
| Consensus | vnGGATTAAndKdKWWhhnn |
| GC content | 36.31% |
| Information content (bits; total / per base) | 15.927 / 0.796 |
| Data sources | HT-SELEX |
| Aligned words | 1787 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (2) | 0.656 | 0.657 | 0.493 | 0.496 | 0.67 | 0.671 | 1.906 | 1.91 | 0.0 | 0 |
| Mouse | 4 (24) | 0.694 | 0.751 | 0.54 | 0.593 | 0.702 | 0.782 | 2.048 | 2.485 | 50.654 | 103.886 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 10 experiments | median | 0.918 | 0.894 | 0.805 | 0.8 | 0.671 | 0.689 |
| best | 0.971 | 0.964 | 0.952 | 0.924 | 0.911 | 0.881 | |
| Methyl HT-SELEX, 2 experiments | median | 0.946 | 0.913 | 0.923 | 0.885 | 0.877 | 0.839 |
| best | 0.968 | 0.946 | 0.952 | 0.924 | 0.911 | 0.881 | |
| Non-Methyl HT-SELEX, 8 experiments | median | 0.909 | 0.894 | 0.755 | 0.753 | 0.636 | 0.655 |
| best | 0.971 | 0.964 | 0.922 | 0.884 | 0.871 | 0.833 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.784 | 0.6 | 0.783 | 0.603 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | Paired-related HD {3.1.3} (TFClass) |
| TF subfamily | PITX {3.1.3.19} (TFClass) |
| TFClass ID | TFClass: 3.1.3.19.1 |
| HGNC | HGNC:9004 |
| MGI | MGI:107374 |
| EntrezGene (human) | GeneID:5307 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:18740 (SSTAR profile) |
| UniProt ID (human) | PITX1_HUMAN |
| UniProt ID (mouse) | PITX1_MOUSE |
| UniProt AC (human) | P78337 (TFClass) |
| UniProt AC (mouse) | P70314 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 4 mouse |
| HT-SELEX | 8 |
| Methyl-HT-SELEX | 2 |
| PCM | PITX1.H12INVIVO.1.S.B.pcm |
| PWM | PITX1.H12INVIVO.1.S.B.pwm |
| PFM | PITX1.H12INVIVO.1.S.B.pfm |
| Alignment | PITX1.H12INVIVO.1.S.B.words.tsv |
| Threshold to P-value map | PITX1.H12INVIVO.1.S.B.thr |
| Motif in other formats | |
| JASPAR format | PITX1.H12INVIVO.1.S.B_jaspar_format.txt |
| MEME format | PITX1.H12INVIVO.1.S.B_meme_format.meme |
| Transfac format | PITX1.H12INVIVO.1.S.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 741.0 | 266.0 | 534.0 | 246.0 |
| 02 | 523.75 | 295.75 | 527.75 | 439.75 |
| 03 | 138.0 | 6.0 | 1561.0 | 82.0 |
| 04 | 74.0 | 1.0 | 1712.0 | 0.0 |
| 05 | 1773.0 | 14.0 | 0.0 | 0.0 |
| 06 | 0.0 | 0.0 | 0.0 | 1787.0 |
| 07 | 0.0 | 70.0 | 0.0 | 1717.0 |
| 08 | 1717.0 | 70.0 | 0.0 | 0.0 |
| 09 | 1626.0 | 30.0 | 109.0 | 22.0 |
| 10 | 324.0 | 466.0 | 606.0 | 391.0 |
| 11 | 318.0 | 223.0 | 977.0 | 269.0 |
| 12 | 195.0 | 195.0 | 1167.0 | 230.0 |
| 13 | 836.0 | 79.0 | 609.0 | 263.0 |
| 14 | 430.0 | 20.0 | 443.0 | 894.0 |
| 15 | 476.0 | 16.0 | 137.0 | 1158.0 |
| 16 | 1012.0 | 40.0 | 16.0 | 719.0 |
| 17 | 893.0 | 186.0 | 147.0 | 561.0 |
| 18 | 654.0 | 444.0 | 193.0 | 496.0 |
| 19 | 502.25 | 593.25 | 261.25 | 430.25 |
| 20 | 360.0 | 660.0 | 302.0 | 465.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.415 | 0.149 | 0.299 | 0.138 |
| 02 | 0.293 | 0.166 | 0.295 | 0.246 |
| 03 | 0.077 | 0.003 | 0.874 | 0.046 |
| 04 | 0.041 | 0.001 | 0.958 | 0.0 |
| 05 | 0.992 | 0.008 | 0.0 | 0.0 |
| 06 | 0.0 | 0.0 | 0.0 | 1.0 |
| 07 | 0.0 | 0.039 | 0.0 | 0.961 |
| 08 | 0.961 | 0.039 | 0.0 | 0.0 |
| 09 | 0.91 | 0.017 | 0.061 | 0.012 |
| 10 | 0.181 | 0.261 | 0.339 | 0.219 |
| 11 | 0.178 | 0.125 | 0.547 | 0.151 |
| 12 | 0.109 | 0.109 | 0.653 | 0.129 |
| 13 | 0.468 | 0.044 | 0.341 | 0.147 |
| 14 | 0.241 | 0.011 | 0.248 | 0.5 |
| 15 | 0.266 | 0.009 | 0.077 | 0.648 |
| 16 | 0.566 | 0.022 | 0.009 | 0.402 |
| 17 | 0.5 | 0.104 | 0.082 | 0.314 |
| 18 | 0.366 | 0.248 | 0.108 | 0.278 |
| 19 | 0.281 | 0.332 | 0.146 | 0.241 |
| 20 | 0.201 | 0.369 | 0.169 | 0.26 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.504 | -0.516 | 0.178 | -0.593 |
| 02 | 0.158 | -0.41 | 0.166 | -0.016 |
| 03 | -1.165 | -4.043 | 1.248 | -1.677 |
| 04 | -1.777 | -5.051 | 1.34 | -5.479 |
| 05 | 1.375 | -3.342 | -5.479 | -5.479 |
| 06 | -5.479 | -5.479 | -5.479 | 1.383 |
| 07 | -5.479 | -1.831 | -5.479 | 1.343 |
| 08 | 1.343 | -1.831 | -5.479 | -5.479 |
| 09 | 1.289 | -2.644 | -1.398 | -2.933 |
| 10 | -0.32 | 0.042 | 0.304 | -0.133 |
| 11 | -0.338 | -0.691 | 0.78 | -0.505 |
| 12 | -0.824 | -0.824 | 0.958 | -0.66 |
| 13 | 0.625 | -1.713 | 0.309 | -0.527 |
| 14 | -0.038 | -3.021 | -0.008 | 0.692 |
| 15 | 0.063 | -3.223 | -1.173 | 0.95 |
| 16 | 0.815 | -2.372 | -3.223 | 0.474 |
| 17 | 0.69 | -0.87 | -1.103 | 0.227 |
| 18 | 0.38 | -0.006 | -0.834 | 0.104 |
| 19 | 0.117 | 0.283 | -0.534 | -0.037 |
| 20 | -0.215 | 0.389 | -0.39 | 0.04 |
| P-value | Threshold |
|---|---|
| 0.001 | 1.85376 |
| 0.0005 | 3.27891 |
| 0.0001 | 6.19246 |