| Motif | PAX8.H12RSNP.0.PSM.A |
| Gene (human) | PAX8 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Pax8 |
| Gene synonyms (mouse) | Pax-8 |
| LOGO |  |
| LOGO (reverse complement) | 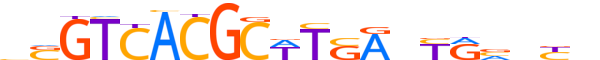 |
| Motif subtype | 0 |
| Quality | A |
| Motif | PAX8.H12RSNP.0.PSM.A |
| Gene (human) | PAX8 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Pax8 |
| Gene synonyms (mouse) | Pax-8 |
| LOGO |  |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 20 |
| Consensus | nRnbYRnTSAWGCGTGACSv |
| GC content | 57.5% |
| Information content (bits; total / per base) | 17.826 / 0.891 |
| Data sources | ChIP-Seq + HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 9993 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (4) | 0.69 | 0.786 | 0.627 | 0.753 | 0.646 | 0.76 | 2.805 | 3.868 | 42.839 | 53.187 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.999 | 0.999 | 0.989 | 0.987 | 0.922 | 0.927 |
| best | 1.0 | 1.0 | 1.0 | 1.0 | 0.999 | 0.999 | |
| Methyl HT-SELEX, 1 experiments | median | 0.999 | 0.999 | 0.977 | 0.974 | 0.846 | 0.855 |
| best | 0.999 | 0.999 | 0.977 | 0.974 | 0.846 | 0.855 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 1.0 | 1.0 | 1.0 | 1.0 | 0.999 | 0.999 |
| best | 1.0 | 1.0 | 1.0 | 1.0 | 0.999 | 0.999 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.718 | 0.44 | 0.73 | 0.474 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Paired box factors {3.2} (TFClass) |
| TF family | PD {3.2.2} (TFClass) |
| TF subfamily | PAX2-like {3.2.2.2} (TFClass) |
| TFClass ID | TFClass: 3.2.2.2.3 |
| HGNC | HGNC:8622 |
| MGI | MGI:97492 |
| EntrezGene (human) | GeneID:7849 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:18510 (SSTAR profile) |
| UniProt ID (human) | PAX8_HUMAN |
| UniProt ID (mouse) | PAX8_MOUSE |
| UniProt AC (human) | Q06710 (TFClass) |
| UniProt AC (mouse) | Q00288 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| PCM | PAX8.H12RSNP.0.PSM.A.pcm |
| PWM | PAX8.H12RSNP.0.PSM.A.pwm |
| PFM | PAX8.H12RSNP.0.PSM.A.pfm |
| Alignment | PAX8.H12RSNP.0.PSM.A.words.tsv |
| Threshold to P-value map | PAX8.H12RSNP.0.PSM.A.thr |
| Motif in other formats | |
| JASPAR format | PAX8.H12RSNP.0.PSM.A_jaspar_format.txt |
| MEME format | PAX8.H12RSNP.0.PSM.A_meme_format.meme |
| Transfac format | PAX8.H12RSNP.0.PSM.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 2089.25 | 1733.25 | 2787.25 | 3383.25 |
| 02 | 3271.25 | 883.25 | 5374.25 | 464.25 |
| 03 | 2553.0 | 2329.0 | 2711.0 | 2400.0 |
| 04 | 127.0 | 2760.0 | 3474.0 | 3632.0 |
| 05 | 483.0 | 6800.0 | 198.0 | 2512.0 |
| 06 | 6944.0 | 614.0 | 2157.0 | 278.0 |
| 07 | 2008.0 | 2599.0 | 3677.0 | 1709.0 |
| 08 | 52.0 | 2009.0 | 175.0 | 7757.0 |
| 09 | 357.0 | 6003.0 | 3509.0 | 124.0 |
| 10 | 8268.0 | 38.0 | 1597.0 | 90.0 |
| 11 | 6187.0 | 522.0 | 175.0 | 3109.0 |
| 12 | 16.0 | 1120.0 | 8814.0 | 43.0 |
| 13 | 42.0 | 9902.0 | 33.0 | 16.0 |
| 14 | 548.0 | 19.0 | 9265.0 | 161.0 |
| 15 | 55.0 | 63.0 | 341.0 | 9534.0 |
| 16 | 1190.0 | 171.0 | 8505.0 | 127.0 |
| 17 | 9178.0 | 141.0 | 437.0 | 237.0 |
| 18 | 569.0 | 9149.0 | 132.0 | 143.0 |
| 19 | 580.0 | 2593.0 | 6112.0 | 708.0 |
| 20 | 2678.5 | 2029.5 | 3963.5 | 1321.5 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.209 | 0.173 | 0.279 | 0.339 |
| 02 | 0.327 | 0.088 | 0.538 | 0.046 |
| 03 | 0.255 | 0.233 | 0.271 | 0.24 |
| 04 | 0.013 | 0.276 | 0.348 | 0.363 |
| 05 | 0.048 | 0.68 | 0.02 | 0.251 |
| 06 | 0.695 | 0.061 | 0.216 | 0.028 |
| 07 | 0.201 | 0.26 | 0.368 | 0.171 |
| 08 | 0.005 | 0.201 | 0.018 | 0.776 |
| 09 | 0.036 | 0.601 | 0.351 | 0.012 |
| 10 | 0.827 | 0.004 | 0.16 | 0.009 |
| 11 | 0.619 | 0.052 | 0.018 | 0.311 |
| 12 | 0.002 | 0.112 | 0.882 | 0.004 |
| 13 | 0.004 | 0.991 | 0.003 | 0.002 |
| 14 | 0.055 | 0.002 | 0.927 | 0.016 |
| 15 | 0.006 | 0.006 | 0.034 | 0.954 |
| 16 | 0.119 | 0.017 | 0.851 | 0.013 |
| 17 | 0.918 | 0.014 | 0.044 | 0.024 |
| 18 | 0.057 | 0.916 | 0.013 | 0.014 |
| 19 | 0.058 | 0.259 | 0.612 | 0.071 |
| 20 | 0.268 | 0.203 | 0.397 | 0.132 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.179 | -0.365 | 0.109 | 0.303 |
| 02 | 0.269 | -1.038 | 0.766 | -1.679 |
| 03 | 0.022 | -0.07 | 0.082 | -0.04 |
| 04 | -2.962 | 0.1 | 0.329 | 0.374 |
| 05 | -1.639 | 1.001 | -2.524 | 0.005 |
| 06 | 1.022 | -1.401 | -0.147 | -2.188 |
| 07 | -0.218 | 0.04 | 0.386 | -0.379 |
| 08 | -3.83 | -0.218 | -2.646 | 1.132 |
| 09 | -1.94 | 0.876 | 0.339 | -2.986 |
| 10 | 1.196 | -4.128 | -0.447 | -3.299 |
| 11 | 0.906 | -1.562 | -2.646 | 0.219 |
| 12 | -4.917 | -0.801 | 1.26 | -4.011 |
| 13 | -4.033 | 1.376 | -4.26 | -4.917 |
| 14 | -1.514 | -4.765 | 1.31 | -2.729 |
| 15 | -3.776 | -3.645 | -1.986 | 1.339 |
| 16 | -0.741 | -2.669 | 1.224 | -2.962 |
| 17 | 1.301 | -2.859 | -1.739 | -2.347 |
| 18 | -1.476 | 1.297 | -2.924 | -2.845 |
| 19 | -1.457 | 0.037 | 0.894 | -1.259 |
| 20 | 0.07 | -0.208 | 0.461 | -0.636 |
| P-value | Threshold |
|---|---|
| 0.001 | 1.33071 |
| 0.0005 | 2.68711 |
| 0.0001 | 5.53181 |