| Motif | NR2E1.H12RSNP.1.S.B |
| Gene (human) | NR2E1 (GeneCards) |
| Gene synonyms (human) | TLX |
| Gene (mouse) | Nr2e1 |
| Gene synonyms (mouse) | Tll, Tlx |
| LOGO | 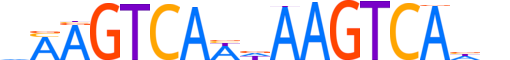 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | B |
| Motif | NR2E1.H12RSNP.1.S.B |
| Gene (human) | NR2E1 (GeneCards) |
| Gene synonyms (human) | TLX |
| Gene (mouse) | Nr2e1 |
| Gene synonyms (mouse) | Tll, Tlx |
| LOGO | 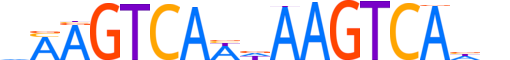 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 17 |
| Consensus | vMAGTCAWdAAGTCAhn |
| GC content | 34.8% |
| Information content (bits; total / per base) | 22.635 / 1.331 |
| Data sources | HT-SELEX |
| Aligned words | 311 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 6 experiments | median | 0.774 | 0.767 | 0.642 | 0.65 | 0.571 | 0.588 |
| best | 0.99 | 0.986 | 0.944 | 0.933 | 0.838 | 0.831 | |
| Methyl HT-SELEX, 1 experiments | median | 0.99 | 0.986 | 0.944 | 0.933 | 0.838 | 0.831 |
| best | 0.99 | 0.986 | 0.944 | 0.933 | 0.838 | 0.831 | |
| Non-Methyl HT-SELEX, 5 experiments | median | 0.735 | 0.727 | 0.621 | 0.627 | 0.561 | 0.575 |
| best | 0.982 | 0.975 | 0.932 | 0.916 | 0.836 | 0.823 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.9 | 0.266 | 0.832 | 0.475 |
| batch 2 | 0.671 | 0.175 | 0.712 | 0.494 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Nuclear receptors with C4 zinc fingers {2.1} (TFClass) |
| TF family | RXR-related receptors {2.1.3} (TFClass) |
| TF subfamily | TLX (NR2E1) {2.1.3.3} (TFClass) |
| TFClass ID | TFClass: 2.1.3.3.1 |
| HGNC | HGNC:7973 |
| MGI | MGI:1100526 |
| EntrezGene (human) | GeneID:7101 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:21907 (SSTAR profile) |
| UniProt ID (human) | NR2E1_HUMAN |
| UniProt ID (mouse) | NR2E1_MOUSE |
| UniProt AC (human) | Q9Y466 (TFClass) |
| UniProt AC (mouse) | Q64104 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 5 |
| Methyl-HT-SELEX | 1 |
| PCM | NR2E1.H12RSNP.1.S.B.pcm |
| PWM | NR2E1.H12RSNP.1.S.B.pwm |
| PFM | NR2E1.H12RSNP.1.S.B.pfm |
| Alignment | NR2E1.H12RSNP.1.S.B.words.tsv |
| Threshold to P-value map | NR2E1.H12RSNP.1.S.B.thr |
| Motif in other formats | |
| JASPAR format | NR2E1.H12RSNP.1.S.B_jaspar_format.txt |
| MEME format | NR2E1.H12RSNP.1.S.B_meme_format.meme |
| Transfac format | NR2E1.H12RSNP.1.S.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 120.0 | 52.0 | 88.0 | 51.0 |
| 02 | 241.75 | 40.75 | 25.75 | 2.75 |
| 03 | 275.0 | 9.0 | 19.0 | 8.0 |
| 04 | 3.0 | 0.0 | 308.0 | 0.0 |
| 05 | 0.0 | 0.0 | 5.0 | 306.0 |
| 06 | 0.0 | 306.0 | 0.0 | 5.0 |
| 07 | 303.0 | 5.0 | 3.0 | 0.0 |
| 08 | 219.0 | 28.0 | 21.0 | 43.0 |
| 09 | 109.0 | 14.0 | 50.0 | 138.0 |
| 10 | 303.0 | 0.0 | 2.0 | 6.0 |
| 11 | 309.0 | 1.0 | 1.0 | 0.0 |
| 12 | 0.0 | 0.0 | 311.0 | 0.0 |
| 13 | 0.0 | 1.0 | 0.0 | 310.0 |
| 14 | 0.0 | 308.0 | 0.0 | 3.0 |
| 15 | 304.0 | 0.0 | 7.0 | 0.0 |
| 16 | 160.0 | 45.0 | 42.0 | 64.0 |
| 17 | 55.75 | 67.75 | 79.75 | 107.75 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.386 | 0.167 | 0.283 | 0.164 |
| 02 | 0.777 | 0.131 | 0.083 | 0.009 |
| 03 | 0.884 | 0.029 | 0.061 | 0.026 |
| 04 | 0.01 | 0.0 | 0.99 | 0.0 |
| 05 | 0.0 | 0.0 | 0.016 | 0.984 |
| 06 | 0.0 | 0.984 | 0.0 | 0.016 |
| 07 | 0.974 | 0.016 | 0.01 | 0.0 |
| 08 | 0.704 | 0.09 | 0.068 | 0.138 |
| 09 | 0.35 | 0.045 | 0.161 | 0.444 |
| 10 | 0.974 | 0.0 | 0.006 | 0.019 |
| 11 | 0.994 | 0.003 | 0.003 | 0.0 |
| 12 | 0.0 | 0.0 | 1.0 | 0.0 |
| 13 | 0.0 | 0.003 | 0.0 | 0.997 |
| 14 | 0.0 | 0.99 | 0.0 | 0.01 |
| 15 | 0.977 | 0.0 | 0.023 | 0.0 |
| 16 | 0.514 | 0.145 | 0.135 | 0.206 |
| 17 | 0.179 | 0.218 | 0.256 | 0.346 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.428 | -0.393 | 0.122 | -0.412 |
| 02 | 1.122 | -0.63 | -1.069 | -2.94 |
| 03 | 1.25 | -2.027 | -1.355 | -2.127 |
| 04 | -2.882 | -4.011 | 1.363 | -4.011 |
| 05 | -4.011 | -4.011 | -2.51 | 1.356 |
| 06 | -4.011 | 1.356 | -4.011 | -2.51 |
| 07 | 1.347 | -2.51 | -2.882 | -4.011 |
| 08 | 1.024 | -0.99 | -1.261 | -0.578 |
| 09 | 0.333 | -1.635 | -0.431 | 0.566 |
| 10 | 1.347 | -4.011 | -3.138 | -2.366 |
| 11 | 1.366 | -3.482 | -3.482 | -4.011 |
| 12 | -4.011 | -4.011 | 1.373 | -4.011 |
| 13 | -4.011 | -3.482 | -4.011 | 1.369 |
| 14 | -4.011 | 1.363 | -4.011 | -2.882 |
| 15 | 1.35 | -4.011 | -2.239 | -4.011 |
| 16 | 0.712 | -0.534 | -0.601 | -0.191 |
| 17 | -0.325 | -0.135 | 0.025 | 0.321 |
| P-value | Threshold |
|---|---|
| 0.001 | -1.66529 |
| 0.0005 | -0.05379 |
| 0.0001 | 3.35646 |