| Motif | NR1I3.H12RSNP.0.P.B |
| Gene (human) | NR1I3 (GeneCards) |
| Gene synonyms (human) | CAR |
| Gene (mouse) | Nr1i3 |
| Gene synonyms (mouse) | Car |
| LOGO |  |
| LOGO (reverse complement) | 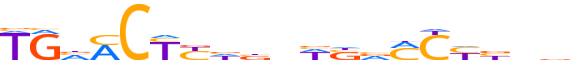 |
| Motif subtype | 0 |
| Quality | B |
| Motif | NR1I3.H12RSNP.0.P.B |
| Gene (human) | NR1I3 (GeneCards) |
| Gene synonyms (human) | CAR |
| Gene (mouse) | Nr1i3 |
| Gene synonyms (mouse) | Car |
| LOGO |  |
| LOGO (reverse complement) | 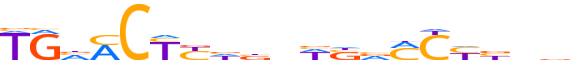 |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 19 |
| Consensus | bhvRGKbSWnvdRRGKbCA |
| GC content | 52.48% |
| Information content (bits; total / per base) | 10.884 / 0.573 |
| Data sources | ChIP-Seq |
| Aligned words | 953 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | 6 (6) | 0.792 | 0.824 | 0.659 | 0.7 | 0.808 | 0.831 | 2.918 | 3.095 | 130.497 | 147.42 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.719 | 0.688 | 0.638 | 0.62 | 0.583 | 0.578 |
| best | 0.77 | 0.734 | 0.678 | 0.654 | 0.611 | 0.602 | |
| Methyl HT-SELEX, 1 experiments | median | 0.668 | 0.641 | 0.598 | 0.586 | 0.555 | 0.554 |
| best | 0.668 | 0.641 | 0.598 | 0.586 | 0.555 | 0.554 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.77 | 0.734 | 0.678 | 0.654 | 0.611 | 0.602 |
| best | 0.77 | 0.734 | 0.678 | 0.654 | 0.611 | 0.602 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.656 | 0.027 | -0.144 | -0.072 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Nuclear receptors with C4 zinc fingers {2.1} (TFClass) |
| TF family | Thyroid hormone receptor-related {2.1.2} (TFClass) |
| TF subfamily | VDR (NR1I) {2.1.2.4} (TFClass) |
| TFClass ID | TFClass: 2.1.2.4.3 |
| HGNC | HGNC:7969 |
| MGI | MGI:1346307 |
| EntrezGene (human) | GeneID:9970 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:12355 (SSTAR profile) |
| UniProt ID (human) | NR1I3_HUMAN |
| UniProt ID (mouse) | NR1I3_MOUSE |
| UniProt AC (human) | Q14994 (TFClass) |
| UniProt AC (mouse) | O35627 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 6 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| PCM | NR1I3.H12RSNP.0.P.B.pcm |
| PWM | NR1I3.H12RSNP.0.P.B.pwm |
| PFM | NR1I3.H12RSNP.0.P.B.pfm |
| Alignment | NR1I3.H12RSNP.0.P.B.words.tsv |
| Threshold to P-value map | NR1I3.H12RSNP.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | NR1I3.H12RSNP.0.P.B_jaspar_format.txt |
| MEME format | NR1I3.H12RSNP.0.P.B_meme_format.meme |
| Transfac format | NR1I3.H12RSNP.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 163.0 | 408.0 | 197.0 | 185.0 |
| 02 | 340.0 | 282.0 | 123.0 | 208.0 |
| 03 | 401.0 | 163.0 | 343.0 | 46.0 |
| 04 | 479.0 | 46.0 | 360.0 | 68.0 |
| 05 | 123.0 | 7.0 | 755.0 | 68.0 |
| 06 | 68.0 | 21.0 | 579.0 | 285.0 |
| 07 | 53.0 | 256.0 | 208.0 | 436.0 |
| 08 | 121.0 | 583.0 | 139.0 | 110.0 |
| 09 | 571.0 | 79.0 | 116.0 | 187.0 |
| 10 | 298.0 | 171.0 | 319.0 | 165.0 |
| 11 | 140.0 | 488.0 | 197.0 | 128.0 |
| 12 | 494.0 | 126.0 | 136.0 | 197.0 |
| 13 | 239.0 | 120.0 | 565.0 | 29.0 |
| 14 | 683.0 | 2.0 | 146.0 | 122.0 |
| 15 | 9.0 | 1.0 | 940.0 | 3.0 |
| 16 | 19.0 | 11.0 | 367.0 | 556.0 |
| 17 | 107.0 | 115.0 | 208.0 | 523.0 |
| 18 | 9.0 | 773.0 | 59.0 | 112.0 |
| 19 | 789.0 | 41.0 | 53.0 | 70.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.171 | 0.428 | 0.207 | 0.194 |
| 02 | 0.357 | 0.296 | 0.129 | 0.218 |
| 03 | 0.421 | 0.171 | 0.36 | 0.048 |
| 04 | 0.503 | 0.048 | 0.378 | 0.071 |
| 05 | 0.129 | 0.007 | 0.792 | 0.071 |
| 06 | 0.071 | 0.022 | 0.608 | 0.299 |
| 07 | 0.056 | 0.269 | 0.218 | 0.458 |
| 08 | 0.127 | 0.612 | 0.146 | 0.115 |
| 09 | 0.599 | 0.083 | 0.122 | 0.196 |
| 10 | 0.313 | 0.179 | 0.335 | 0.173 |
| 11 | 0.147 | 0.512 | 0.207 | 0.134 |
| 12 | 0.518 | 0.132 | 0.143 | 0.207 |
| 13 | 0.251 | 0.126 | 0.593 | 0.03 |
| 14 | 0.717 | 0.002 | 0.153 | 0.128 |
| 15 | 0.009 | 0.001 | 0.986 | 0.003 |
| 16 | 0.02 | 0.012 | 0.385 | 0.583 |
| 17 | 0.112 | 0.121 | 0.218 | 0.549 |
| 18 | 0.009 | 0.811 | 0.062 | 0.118 |
| 19 | 0.828 | 0.043 | 0.056 | 0.073 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.376 | 0.535 | -0.189 | -0.251 |
| 02 | 0.353 | 0.167 | -0.654 | -0.135 |
| 03 | 0.518 | -0.376 | 0.362 | -1.615 |
| 04 | 0.695 | -1.615 | 0.41 | -1.236 |
| 05 | -0.654 | -3.315 | 1.148 | -1.236 |
| 06 | -1.236 | -2.357 | 0.884 | 0.178 |
| 07 | -1.478 | 0.071 | -0.135 | 0.601 |
| 08 | -0.671 | 0.891 | -0.534 | -0.765 |
| 09 | 0.87 | -1.09 | -0.712 | -0.24 |
| 10 | 0.222 | -0.329 | 0.29 | -0.364 |
| 11 | -0.527 | 0.713 | -0.189 | -0.615 |
| 12 | 0.726 | -0.631 | -0.555 | -0.189 |
| 13 | 0.003 | -0.679 | 0.859 | -2.056 |
| 14 | 1.049 | -4.168 | -0.485 | -0.663 |
| 15 | -3.109 | -4.482 | 1.367 | -3.93 |
| 16 | -2.45 | -2.938 | 0.43 | 0.843 |
| 17 | -0.792 | -0.721 | -0.135 | 0.782 |
| 18 | -3.109 | 1.172 | -1.374 | -0.747 |
| 19 | 1.192 | -1.726 | -1.478 | -1.208 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.56306 |
| 0.0005 | 5.41726 |
| 0.0001 | 7.17286 |