| Motif | NPAS2.H12CORE.0.M.B |
| Gene (human) | NPAS2 (GeneCards) |
| Gene synonyms (human) | BHLHE9, MOP4, PASD4 |
| Gene (mouse) | Npas2 |
| Gene synonyms (mouse) | |
| LOGO | 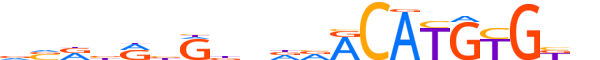 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | NPAS2.H12CORE.0.M.B |
| Gene (human) | NPAS2 (GeneCards) |
| Gene synonyms (human) | BHLHE9, MOP4, PASD4 |
| Gene (mouse) | Npas2 |
| Gene synonyms (mouse) | |
| LOGO | 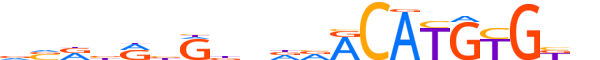 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 20 |
| Consensus | vvRbRbRbndvACATGTGKn |
| GC content | 51.48% |
| Information content (bits; total / per base) | 14.55 / 0.727 |
| Data sources | Methyl-HT-SELEX |
| Aligned words | 379 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.982 | 0.977 | 0.911 | 0.903 | 0.818 | 0.816 |
| best | 0.998 | 0.997 | 0.994 | 0.991 | 0.984 | 0.978 | |
| Methyl HT-SELEX, 2 experiments | median | 0.857 | 0.852 | 0.797 | 0.791 | 0.747 | 0.741 |
| best | 0.988 | 0.983 | 0.969 | 0.957 | 0.921 | 0.903 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.987 | 0.984 | 0.923 | 0.92 | 0.849 | 0.853 |
| best | 0.998 | 0.997 | 0.994 | 0.991 | 0.984 | 0.978 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.711 | 0.551 | 0.548 | 0.326 |
| TF superclass | Basic domains {1} (TFClass) |
| TF class | Basic helix-loop-helix factors (bHLH) {1.2} (TFClass) |
| TF family | PAS {1.2.5} (TFClass) |
| TF subfamily | PAS-ARNT {1.2.5.2} (TFClass) |
| TFClass ID | TFClass: 1.2.5.2.6 |
| HGNC | HGNC:7895 |
| MGI | MGI:109232 |
| EntrezGene (human) | GeneID:4862 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:18143 (SSTAR profile) |
| UniProt ID (human) | NPAS2_HUMAN |
| UniProt ID (mouse) | NPAS2_MOUSE |
| UniProt AC (human) | Q99743 (TFClass) |
| UniProt AC (mouse) | P97460 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 2 |
| PCM | NPAS2.H12CORE.0.M.B.pcm |
| PWM | NPAS2.H12CORE.0.M.B.pwm |
| PFM | NPAS2.H12CORE.0.M.B.pfm |
| Alignment | NPAS2.H12CORE.0.M.B.words.tsv |
| Threshold to P-value map | NPAS2.H12CORE.0.M.B.thr |
| Motif in other formats | |
| JASPAR format | NPAS2.H12CORE.0.M.B_jaspar_format.txt |
| MEME format | NPAS2.H12CORE.0.M.B_meme_format.meme |
| Transfac format | NPAS2.H12CORE.0.M.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 154.75 | 62.75 | 118.75 | 42.75 |
| 02 | 69.5 | 209.5 | 75.5 | 24.5 |
| 03 | 194.0 | 21.0 | 134.0 | 30.0 |
| 04 | 39.0 | 101.0 | 76.0 | 163.0 |
| 05 | 124.0 | 14.0 | 216.0 | 25.0 |
| 06 | 37.0 | 80.0 | 93.0 | 169.0 |
| 07 | 35.0 | 17.0 | 294.0 | 33.0 |
| 08 | 45.0 | 49.0 | 145.0 | 140.0 |
| 09 | 143.0 | 63.0 | 97.0 | 76.0 |
| 10 | 190.0 | 17.0 | 110.0 | 62.0 |
| 11 | 208.0 | 49.0 | 97.0 | 25.0 |
| 12 | 284.0 | 19.0 | 76.0 | 0.0 |
| 13 | 1.0 | 376.0 | 2.0 | 0.0 |
| 14 | 357.0 | 0.0 | 22.0 | 0.0 |
| 15 | 0.0 | 72.0 | 0.0 | 307.0 |
| 16 | 48.0 | 0.0 | 331.0 | 0.0 |
| 17 | 1.0 | 128.0 | 0.0 | 250.0 |
| 18 | 2.0 | 0.0 | 374.0 | 3.0 |
| 19 | 7.0 | 74.0 | 97.0 | 201.0 |
| 20 | 77.5 | 103.5 | 88.5 | 109.5 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.408 | 0.166 | 0.313 | 0.113 |
| 02 | 0.183 | 0.553 | 0.199 | 0.065 |
| 03 | 0.512 | 0.055 | 0.354 | 0.079 |
| 04 | 0.103 | 0.266 | 0.201 | 0.43 |
| 05 | 0.327 | 0.037 | 0.57 | 0.066 |
| 06 | 0.098 | 0.211 | 0.245 | 0.446 |
| 07 | 0.092 | 0.045 | 0.776 | 0.087 |
| 08 | 0.119 | 0.129 | 0.383 | 0.369 |
| 09 | 0.377 | 0.166 | 0.256 | 0.201 |
| 10 | 0.501 | 0.045 | 0.29 | 0.164 |
| 11 | 0.549 | 0.129 | 0.256 | 0.066 |
| 12 | 0.749 | 0.05 | 0.201 | 0.0 |
| 13 | 0.003 | 0.992 | 0.005 | 0.0 |
| 14 | 0.942 | 0.0 | 0.058 | 0.0 |
| 15 | 0.0 | 0.19 | 0.0 | 0.81 |
| 16 | 0.127 | 0.0 | 0.873 | 0.0 |
| 17 | 0.003 | 0.338 | 0.0 | 0.66 |
| 18 | 0.005 | 0.0 | 0.987 | 0.008 |
| 19 | 0.018 | 0.195 | 0.256 | 0.53 |
| 20 | 0.204 | 0.273 | 0.234 | 0.289 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.485 | -0.404 | 0.223 | -0.777 |
| 02 | -0.304 | 0.785 | -0.223 | -1.309 |
| 03 | 0.709 | -1.454 | 0.342 | -1.117 |
| 04 | -0.866 | 0.063 | -0.217 | 0.536 |
| 05 | 0.265 | -1.827 | 0.815 | -1.29 |
| 06 | -0.917 | -0.166 | -0.018 | 0.572 |
| 07 | -0.97 | -1.65 | 1.122 | -1.026 |
| 08 | -0.728 | -0.645 | 0.42 | 0.385 |
| 09 | 0.406 | -0.4 | 0.023 | -0.217 |
| 10 | 0.688 | -1.65 | 0.147 | -0.416 |
| 11 | 0.778 | -0.645 | 0.023 | -1.29 |
| 12 | 1.087 | -1.547 | -0.217 | -4.172 |
| 13 | -3.657 | 1.367 | -3.318 | -4.172 |
| 14 | 1.315 | -4.172 | -1.41 | -4.172 |
| 15 | -4.172 | -0.27 | -4.172 | 1.165 |
| 16 | -0.665 | -4.172 | 1.24 | -4.172 |
| 17 | -3.657 | 0.297 | -4.172 | 0.961 |
| 18 | -3.318 | -4.172 | 1.361 | -3.066 |
| 19 | -2.429 | -0.243 | 0.023 | 0.744 |
| 20 | -0.198 | 0.087 | -0.067 | 0.143 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.26876 |
| 0.0005 | 4.44246 |
| 0.0001 | 6.85211 |