| Motif | NKX32.H12RSNP.0.PSM.D |
| Gene (human) | NKX3-2 (GeneCards) |
| Gene synonyms (human) | BAPX1, NKX3B |
| Gene (mouse) | Nkx3-2 |
| Gene synonyms (mouse) | Bapx1, Nkx-3.2, Nkx3b |
| LOGO | 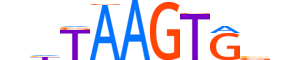 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | D |
| Motif | NKX32.H12RSNP.0.PSM.D |
| Gene (human) | NKX3-2 (GeneCards) |
| Gene synonyms (human) | BAPX1, NKX3B |
| Gene (mouse) | Nkx3-2 |
| Gene synonyms (mouse) | Bapx1, Nkx-3.2, Nkx3b |
| LOGO | 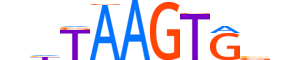 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | D |
| Motif length | 10 |
| Consensus | nhTAAGTGbn |
| GC content | 41.01% |
| Information content (bits; total / per base) | 10.198 / 1.02 |
| Data sources | ChIP-Seq + HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 9976 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | 2 (13) | 0.863 | 0.975 | 0.802 | 0.949 | 0.794 | 0.929 | 2.48 | 3.372 | 229.292 | 419.237 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.976 | 0.963 | 0.921 | 0.904 | 0.802 | 0.808 |
| best | 0.994 | 0.991 | 0.978 | 0.971 | 0.894 | 0.873 | |
| Methyl HT-SELEX, 1 experiments | median | 0.93 | 0.897 | 0.848 | 0.816 | 0.726 | 0.719 |
| best | 0.93 | 0.897 | 0.848 | 0.816 | 0.726 | 0.719 | |
| Non-Methyl HT-SELEX, 3 experiments | median | 0.981 | 0.97 | 0.957 | 0.94 | 0.864 | 0.868 |
| best | 0.994 | 0.991 | 0.978 | 0.971 | 0.894 | 0.873 | |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | NK-related {3.1.2} (TFClass) |
| TF subfamily | NK3 {3.1.2.16} (TFClass) |
| TFClass ID | TFClass: 3.1.2.16.2 |
| HGNC | HGNC:951 |
| MGI | MGI:108015 |
| EntrezGene (human) | GeneID:579 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:12020 (SSTAR profile) |
| UniProt ID (human) | NKX32_HUMAN |
| UniProt ID (mouse) | NKX32_MOUSE |
| UniProt AC (human) | P78367 (TFClass) |
| UniProt AC (mouse) | P97503 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 2 mouse |
| HT-SELEX | 3 |
| Methyl-HT-SELEX | 1 |
| PCM | NKX32.H12RSNP.0.PSM.D.pcm |
| PWM | NKX32.H12RSNP.0.PSM.D.pwm |
| PFM | NKX32.H12RSNP.0.PSM.D.pfm |
| Alignment | NKX32.H12RSNP.0.PSM.D.words.tsv |
| Threshold to P-value map | NKX32.H12RSNP.0.PSM.D.thr |
| Motif in other formats | |
| JASPAR format | NKX32.H12RSNP.0.PSM.D_jaspar_format.txt |
| MEME format | NKX32.H12RSNP.0.PSM.D_meme_format.meme |
| Transfac format | NKX32.H12RSNP.0.PSM.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 2316.5 | 2585.5 | 3321.5 | 1752.5 |
| 02 | 1649.25 | 1915.25 | 1445.25 | 4966.25 |
| 03 | 407.0 | 407.0 | 643.0 | 8519.0 |
| 04 | 9783.0 | 0.0 | 181.0 | 12.0 |
| 05 | 9976.0 | 0.0 | 0.0 | 0.0 |
| 06 | 4.0 | 8.0 | 9918.0 | 46.0 |
| 07 | 195.0 | 56.0 | 276.0 | 9449.0 |
| 08 | 1906.0 | 89.0 | 7873.0 | 108.0 |
| 09 | 932.75 | 3272.75 | 3964.75 | 1805.75 |
| 10 | 1809.0 | 2526.0 | 2425.0 | 3216.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.232 | 0.259 | 0.333 | 0.176 |
| 02 | 0.165 | 0.192 | 0.145 | 0.498 |
| 03 | 0.041 | 0.041 | 0.064 | 0.854 |
| 04 | 0.981 | 0.0 | 0.018 | 0.001 |
| 05 | 1.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 0.001 | 0.994 | 0.005 |
| 07 | 0.02 | 0.006 | 0.028 | 0.947 |
| 08 | 0.191 | 0.009 | 0.789 | 0.011 |
| 09 | 0.093 | 0.328 | 0.397 | 0.181 |
| 10 | 0.181 | 0.253 | 0.243 | 0.322 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.074 | 0.036 | 0.286 | -0.352 |
| 02 | -0.413 | -0.264 | -0.545 | 0.688 |
| 03 | -1.808 | -1.808 | -1.353 | 1.228 |
| 04 | 1.366 | -6.989 | -2.611 | -5.162 |
| 05 | 1.386 | -6.989 | -6.989 | -6.989 |
| 06 | -5.982 | -5.49 | 1.38 | -3.945 |
| 07 | -2.538 | -3.757 | -2.194 | 1.331 |
| 08 | -0.269 | -3.308 | 1.149 | -3.119 |
| 09 | -0.982 | 0.272 | 0.463 | -0.323 |
| 10 | -0.321 | 0.013 | -0.028 | 0.254 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.637045 |
| 0.0005 | 5.685825 |
| 0.0001 | 7.75514 |