| Motif | NKX32.H12INVIVO.1.S.B |
| Gene (human) | NKX3-2 (GeneCards) |
| Gene synonyms (human) | BAPX1, NKX3B |
| Gene (mouse) | Nkx3-2 |
| Gene synonyms (mouse) | Bapx1, Nkx-3.2, Nkx3b |
| LOGO | 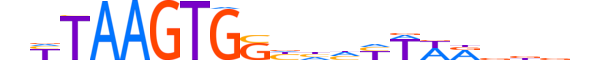 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | B |
| Motif | NKX32.H12INVIVO.1.S.B |
| Gene (human) | NKX3-2 (GeneCards) |
| Gene synonyms (human) | BAPX1, NKX3B |
| Gene (mouse) | Nkx3-2 |
| Gene synonyms (mouse) | Bapx1, Nkx-3.2, Nkx3b |
| LOGO | 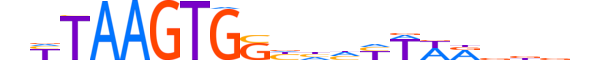 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 20 |
| Consensus | nYTAAGTGShhhYWWRdddn |
| GC content | 41.03% |
| Information content (bits; total / per base) | 16.767 / 0.838 |
| Data sources | HT-SELEX |
| Aligned words | 1560 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | 2 (13) | 0.835 | 0.954 | 0.764 | 0.908 | 0.761 | 0.915 | 2.526 | 3.858 | 197.638 | 375.292 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.949 | 0.931 | 0.862 | 0.843 | 0.735 | 0.741 |
| best | 0.993 | 0.991 | 0.938 | 0.932 | 0.851 | 0.823 | |
| Methyl HT-SELEX, 1 experiments | median | 0.894 | 0.857 | 0.798 | 0.766 | 0.69 | 0.681 |
| best | 0.894 | 0.857 | 0.798 | 0.766 | 0.69 | 0.681 | |
| Non-Methyl HT-SELEX, 3 experiments | median | 0.961 | 0.942 | 0.922 | 0.893 | 0.78 | 0.797 |
| best | 0.993 | 0.991 | 0.938 | 0.932 | 0.851 | 0.823 | |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | NK-related {3.1.2} (TFClass) |
| TF subfamily | NK3 {3.1.2.16} (TFClass) |
| TFClass ID | TFClass: 3.1.2.16.2 |
| HGNC | HGNC:951 |
| MGI | MGI:108015 |
| EntrezGene (human) | GeneID:579 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:12020 (SSTAR profile) |
| UniProt ID (human) | NKX32_HUMAN |
| UniProt ID (mouse) | NKX32_MOUSE |
| UniProt AC (human) | P78367 (TFClass) |
| UniProt AC (mouse) | P97503 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 2 mouse |
| HT-SELEX | 3 |
| Methyl-HT-SELEX | 1 |
| PCM | NKX32.H12INVIVO.1.S.B.pcm |
| PWM | NKX32.H12INVIVO.1.S.B.pwm |
| PFM | NKX32.H12INVIVO.1.S.B.pfm |
| Alignment | NKX32.H12INVIVO.1.S.B.words.tsv |
| Threshold to P-value map | NKX32.H12INVIVO.1.S.B.thr |
| Motif in other formats | |
| JASPAR format | NKX32.H12INVIVO.1.S.B_jaspar_format.txt |
| MEME format | NKX32.H12INVIVO.1.S.B_meme_format.meme |
| Transfac format | NKX32.H12INVIVO.1.S.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 280.25 | 605.25 | 339.25 | 335.25 |
| 02 | 186.0 | 249.0 | 101.0 | 1024.0 |
| 03 | 24.0 | 44.0 | 31.0 | 1461.0 |
| 04 | 1553.0 | 0.0 | 7.0 | 0.0 |
| 05 | 1559.0 | 0.0 | 0.0 | 1.0 |
| 06 | 0.0 | 0.0 | 1559.0 | 1.0 |
| 07 | 3.0 | 0.0 | 3.0 | 1554.0 |
| 08 | 57.0 | 0.0 | 1503.0 | 0.0 |
| 09 | 13.0 | 456.0 | 1034.0 | 57.0 |
| 10 | 174.0 | 876.0 | 156.0 | 354.0 |
| 11 | 651.0 | 502.0 | 73.0 | 334.0 |
| 12 | 555.0 | 695.0 | 76.0 | 234.0 |
| 13 | 273.0 | 385.0 | 23.0 | 879.0 |
| 14 | 226.0 | 32.0 | 108.0 | 1194.0 |
| 15 | 929.0 | 39.0 | 81.0 | 511.0 |
| 16 | 955.0 | 56.0 | 319.0 | 230.0 |
| 17 | 314.0 | 201.0 | 681.0 | 364.0 |
| 18 | 249.0 | 176.0 | 561.0 | 574.0 |
| 19 | 322.75 | 247.75 | 697.75 | 291.75 |
| 20 | 387.5 | 351.5 | 532.5 | 288.5 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.18 | 0.388 | 0.217 | 0.215 |
| 02 | 0.119 | 0.16 | 0.065 | 0.656 |
| 03 | 0.015 | 0.028 | 0.02 | 0.937 |
| 04 | 0.996 | 0.0 | 0.004 | 0.0 |
| 05 | 0.999 | 0.0 | 0.0 | 0.001 |
| 06 | 0.0 | 0.0 | 0.999 | 0.001 |
| 07 | 0.002 | 0.0 | 0.002 | 0.996 |
| 08 | 0.037 | 0.0 | 0.963 | 0.0 |
| 09 | 0.008 | 0.292 | 0.663 | 0.037 |
| 10 | 0.112 | 0.562 | 0.1 | 0.227 |
| 11 | 0.417 | 0.322 | 0.047 | 0.214 |
| 12 | 0.356 | 0.446 | 0.049 | 0.15 |
| 13 | 0.175 | 0.247 | 0.015 | 0.563 |
| 14 | 0.145 | 0.021 | 0.069 | 0.765 |
| 15 | 0.596 | 0.025 | 0.052 | 0.328 |
| 16 | 0.612 | 0.036 | 0.204 | 0.147 |
| 17 | 0.201 | 0.129 | 0.437 | 0.233 |
| 18 | 0.16 | 0.113 | 0.36 | 0.368 |
| 19 | 0.207 | 0.159 | 0.447 | 0.187 |
| 20 | 0.248 | 0.225 | 0.341 | 0.185 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.329 | 0.438 | -0.139 | -0.151 |
| 02 | -0.735 | -0.446 | -1.338 | 0.962 |
| 03 | -2.719 | -2.146 | -2.479 | 1.317 |
| 04 | 1.378 | -5.362 | -3.792 | -5.362 |
| 05 | 1.382 | -5.362 | -5.362 | -4.928 |
| 06 | -5.362 | -5.362 | 1.382 | -4.928 |
| 07 | -4.394 | -5.362 | -4.394 | 1.379 |
| 08 | -1.896 | -5.362 | 1.346 | -5.362 |
| 09 | -3.274 | 0.156 | 0.972 | -1.896 |
| 10 | -0.801 | 0.807 | -0.909 | -0.096 |
| 11 | 0.51 | 0.251 | -1.656 | -0.154 |
| 12 | 0.351 | 0.576 | -1.616 | -0.508 |
| 13 | -0.355 | -0.013 | -2.758 | 0.81 |
| 14 | -0.542 | -2.449 | -1.272 | 1.116 |
| 15 | 0.865 | -2.261 | -1.554 | 0.269 |
| 16 | 0.893 | -1.913 | -0.2 | -0.525 |
| 17 | -0.216 | -0.658 | 0.555 | -0.069 |
| 18 | -0.446 | -0.79 | 0.362 | 0.385 |
| 19 | -0.188 | -0.451 | 0.58 | -0.289 |
| 20 | -0.006 | -0.103 | 0.31 | -0.3 |
| P-value | Threshold |
|---|---|
| 0.001 | 1.23306 |
| 0.0005 | 2.72656 |
| 0.0001 | 5.81651 |