| Motif | NKX25.H12INVIVO.0.PSM.A |
| Gene (human) | NKX2-5 (GeneCards) |
| Gene synonyms (human) | CSX, NKX2.5, NKX2E |
| Gene (mouse) | Nkx2-5 |
| Gene synonyms (mouse) | Csx, Nkx-2.5, Nkx2e |
| LOGO | 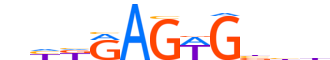 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | A |
| Motif | NKX25.H12INVIVO.0.PSM.A |
| Gene (human) | NKX2-5 (GeneCards) |
| Gene synonyms (human) | CSX, NKX2.5, NKX2E |
| Gene (mouse) | Nkx2-5 |
| Gene synonyms (mouse) | Csx, Nkx-2.5, Nkx2e |
| LOGO | 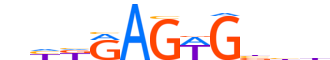 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 11 |
| Consensus | nhKRAGWGbbh |
| GC content | 50.76% |
| Information content (bits; total / per base) | 7.986 / 0.726 |
| Data sources | ChIP-Seq + HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 989 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 2 (14) | 0.856 | 0.865 | 0.726 | 0.741 | 0.742 | 0.753 | 1.944 | 2.003 | 166.214 | 196.276 |
| Mouse | 7 (29) | 0.765 | 0.91 | 0.609 | 0.81 | 0.653 | 0.818 | 1.53 | 2.407 | 68.824 | 151.328 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.884 | 0.817 | 0.854 | 0.79 | 0.794 | 0.743 |
| best | 0.898 | 0.835 | 0.873 | 0.811 | 0.819 | 0.767 | |
| Methyl HT-SELEX, 1 experiments | median | 0.898 | 0.835 | 0.873 | 0.811 | 0.819 | 0.767 |
| best | 0.898 | 0.835 | 0.873 | 0.811 | 0.819 | 0.767 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.869 | 0.798 | 0.835 | 0.768 | 0.769 | 0.72 |
| best | 0.869 | 0.798 | 0.835 | 0.768 | 0.769 | 0.72 | |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | NK-related {3.1.2} (TFClass) |
| TF subfamily | NK4 {3.1.2.17} (TFClass) |
| TFClass ID | TFClass: 3.1.2.17.2 |
| HGNC | HGNC:2488 |
| MGI | MGI:97350 |
| EntrezGene (human) | GeneID:1482 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:18091 (SSTAR profile) |
| UniProt ID (human) | NKX25_HUMAN |
| UniProt ID (mouse) | NKX25_MOUSE |
| UniProt AC (human) | P52952 (TFClass) |
| UniProt AC (mouse) | P42582 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 7 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| PCM | NKX25.H12INVIVO.0.PSM.A.pcm |
| PWM | NKX25.H12INVIVO.0.PSM.A.pwm |
| PFM | NKX25.H12INVIVO.0.PSM.A.pfm |
| Alignment | NKX25.H12INVIVO.0.PSM.A.words.tsv |
| Threshold to P-value map | NKX25.H12INVIVO.0.PSM.A.thr |
| Motif in other formats | |
| JASPAR format | NKX25.H12INVIVO.0.PSM.A_jaspar_format.txt |
| MEME format | NKX25.H12INVIVO.0.PSM.A_meme_format.meme |
| Transfac format | NKX25.H12INVIVO.0.PSM.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 122.0 | 309.0 | 303.0 | 255.0 |
| 02 | 181.0 | 233.0 | 49.0 | 526.0 |
| 03 | 116.0 | 81.0 | 210.0 | 582.0 |
| 04 | 210.0 | 61.0 | 715.0 | 3.0 |
| 05 | 976.0 | 2.0 | 7.0 | 4.0 |
| 06 | 9.0 | 63.0 | 906.0 | 11.0 |
| 07 | 317.0 | 4.0 | 53.0 | 615.0 |
| 08 | 62.0 | 9.0 | 917.0 | 1.0 |
| 09 | 68.0 | 286.0 | 404.0 | 231.0 |
| 10 | 100.0 | 321.0 | 230.0 | 338.0 |
| 11 | 174.0 | 205.0 | 154.0 | 456.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.123 | 0.312 | 0.306 | 0.258 |
| 02 | 0.183 | 0.236 | 0.05 | 0.532 |
| 03 | 0.117 | 0.082 | 0.212 | 0.588 |
| 04 | 0.212 | 0.062 | 0.723 | 0.003 |
| 05 | 0.987 | 0.002 | 0.007 | 0.004 |
| 06 | 0.009 | 0.064 | 0.916 | 0.011 |
| 07 | 0.321 | 0.004 | 0.054 | 0.622 |
| 08 | 0.063 | 0.009 | 0.927 | 0.001 |
| 09 | 0.069 | 0.289 | 0.408 | 0.234 |
| 10 | 0.101 | 0.325 | 0.233 | 0.342 |
| 11 | 0.176 | 0.207 | 0.156 | 0.461 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.699 | 0.222 | 0.202 | 0.031 |
| 02 | -0.309 | -0.059 | -1.591 | 0.751 |
| 03 | -0.749 | -1.102 | -0.162 | 0.852 |
| 04 | -0.162 | -1.379 | 1.057 | -3.965 |
| 05 | 1.368 | -4.203 | -3.351 | -3.773 |
| 06 | -3.145 | -1.347 | 1.294 | -2.974 |
| 07 | 0.247 | -3.773 | -1.515 | 0.907 |
| 08 | -1.363 | -3.145 | 1.306 | -4.515 |
| 09 | -1.273 | 0.145 | 0.488 | -0.067 |
| 10 | -0.895 | 0.259 | -0.072 | 0.311 |
| 11 | -0.348 | -0.186 | -0.469 | 0.609 |
| P-value | Threshold |
|---|---|
| 0.001 | 5.238155 |
| 0.0005 | 5.898945 |
| 0.0001 | 7.10379 |